Proteins (6)
id: wild-bear-maple
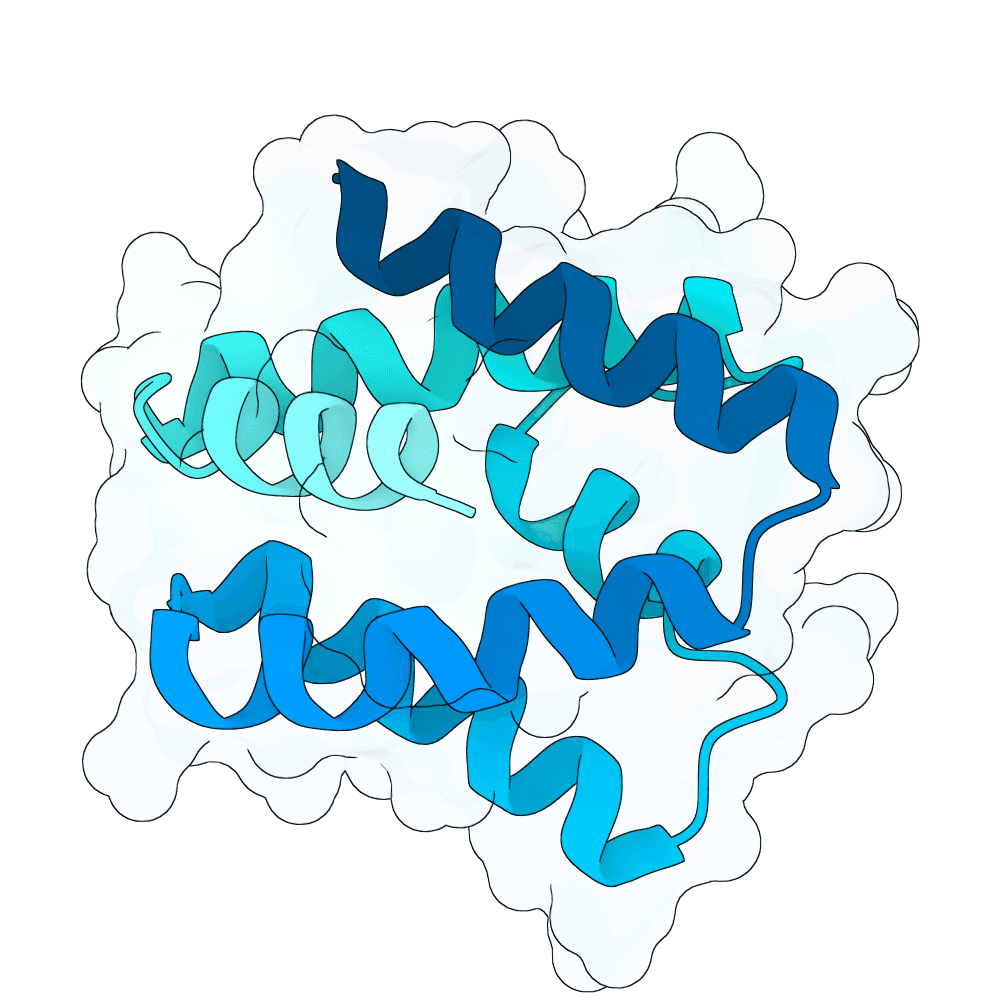
Nipah Virus Glycoprotein G
None
89.38
True
12.5 kDa
109
id: crimson-falcon-orchid
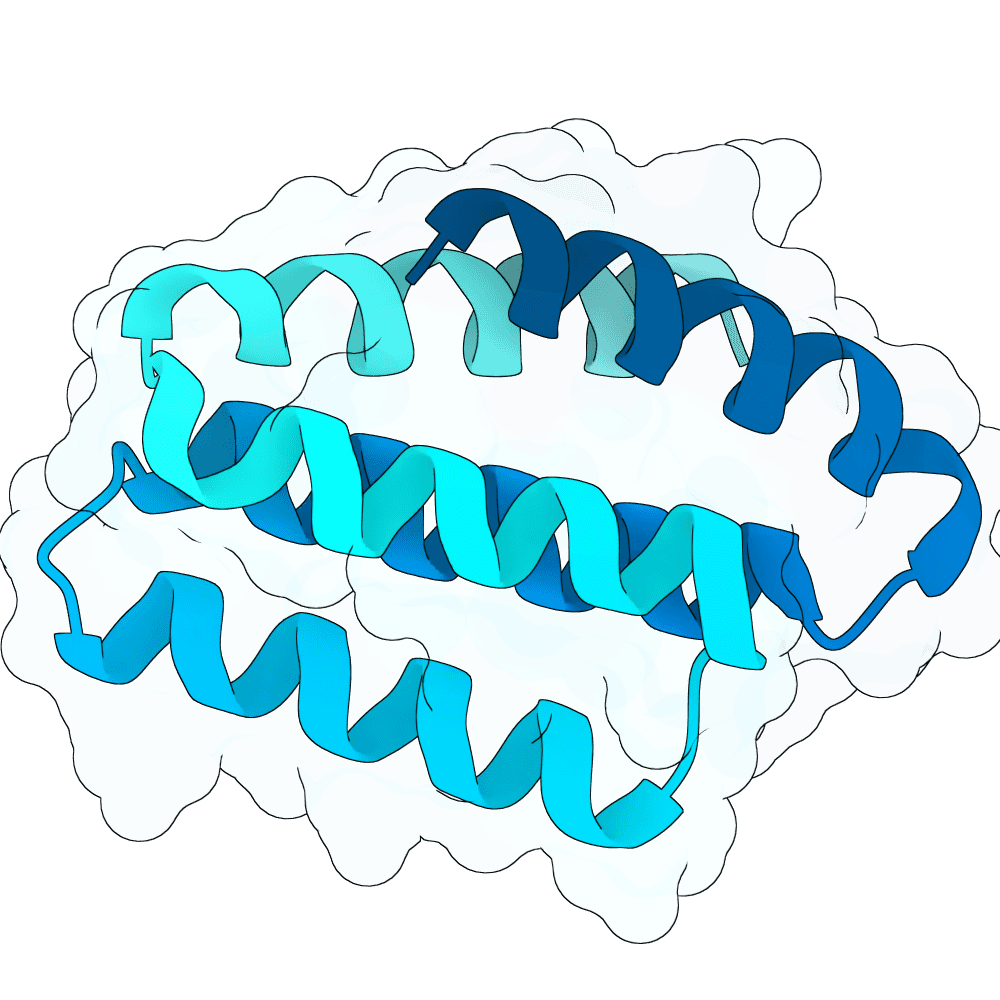
Nipah Virus Glycoprotein G
None
89.07
True
10.7 kDa
96
id: pale-bear-thorn
Nipah Virus Glycoprotein G
None
60.22
True
13.2 kDa
116
id: shy-bison-sand
Nipah Virus Glycoprotein G
None
86.44
True
7.0 kDa
58
id: strong-tiger-thorn

Nipah Virus Glycoprotein G
None
68.23
True
6.8 kDa
58
id: green-bear-cedar
Nipah Virus Glycoprotein G
0.78
88.45
--
10.6 kDa
86