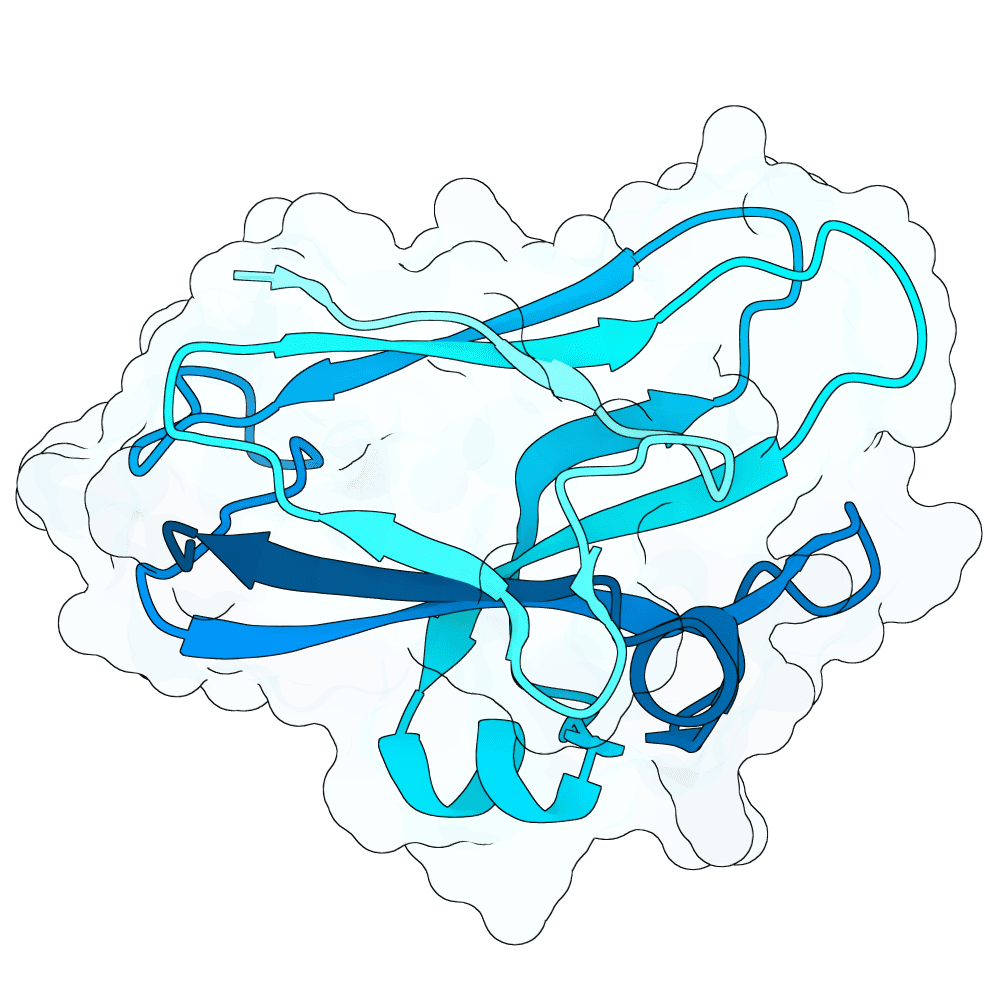
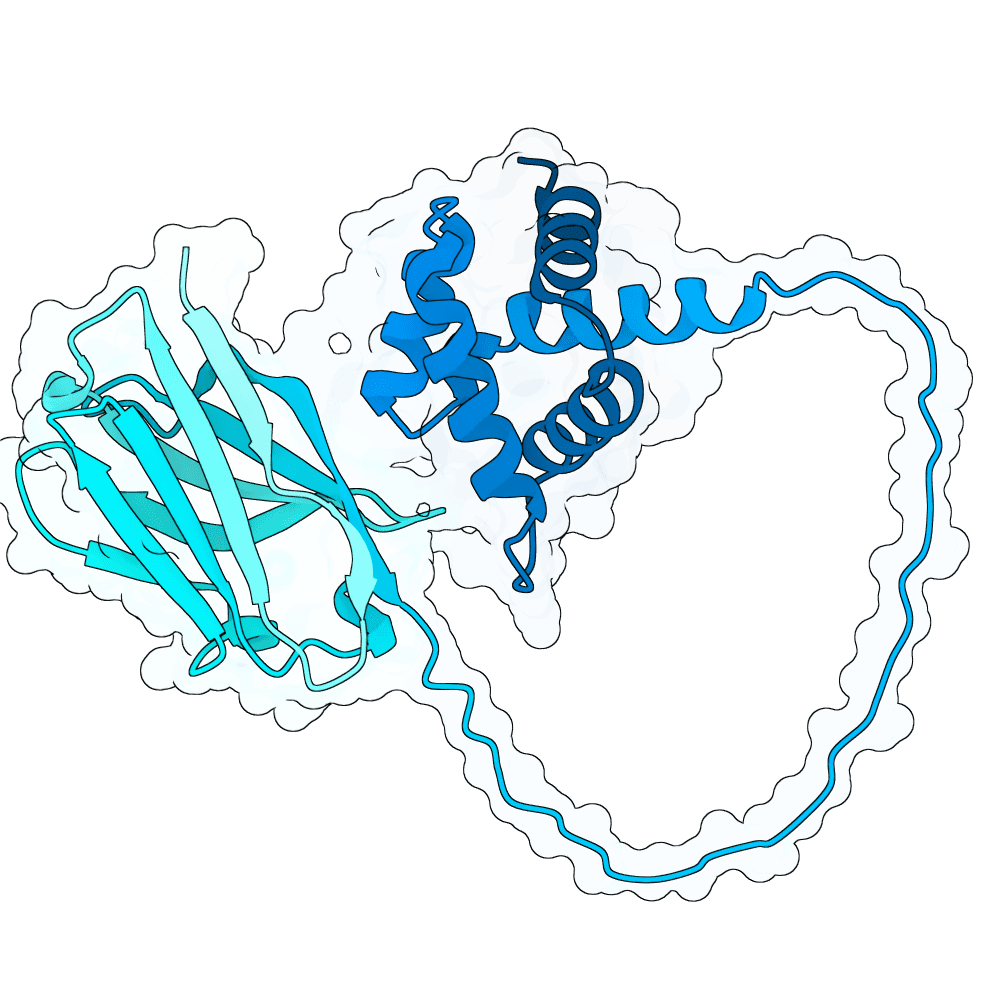
Description
This collection contains experimental validation results from the Adaptyv Nipah Binder Competition, an open challenge that invited protein designers from around the world to design binders against Nipah virus, a WHO priority pathogen with 40-75% fatality rates and no approved treatments. The goal: accelerate therapeutic development while generating large-scale data to benchmark computational methods and train AI models. 680+ participants submitted over 10,000 designs targeting Nipah Glycoprotein G, a challenging tetramer with limited structural data and little prior binder work to build on. 1,200 were selected via Boltz-2 ipSAE ranking, community voting, and expert curation, then validated using Adaptyv's standardized expression and binding affinity characterization pipeline.
Results Overview
This release includes characterization data for 1,026 designs plus 4 positive controls. The remaining 174 designs are still undergoing characterization and we expect to share this data within the next few days. Preliminary analysis suggests these outstanding samples fall between non-binders and weak binders, with unclear signal making characterization more difficult. We don't expect the top rankings to change significantly once this data is added.
Out of 1,026 designs tested, 881 expressed successfully (86%) and 99 showed binding (9.6%). 26 designs achieved single-digit nanomolar affinity or better.
General Ranking: Top 3 Best Binders:
- 🥇 Miles McGibbon (KD: 3.7e-10 M)
- 🥈 Liviu Copoiu (KD: 4.5e-10 M)
- 🥉 Andrew Huang (KD: 7.2e-10 M)
De Novo Ranking: Top 3 Best Binders:
- 🥇 Nick Boyd (KD: 1.4e-9 M)
- 🥈 Johannes Klier (KD: 1.2e-8 M)
- 🥉 Brian Coventry (KD: 1.8e-8 M)
Beyond binding affinity
We ran additional experiments to assess properties relevant to therapeutic development. Neutralization data for top binders is included in this release; HSA off-target binding and expression yield data for all designs are still being processed and will be released in the next few days.
- Neutralization: Can the binder displace the natural ephrin-B2/B3 receptor? This is a key indicator of whether a design could actually block viral entry. Top binders are tested using a competitive BLI assay where the variant competes against ephrin-B2 for binding.
- HSA off-target binding: Human Serum Albumin is abundant in the body, so any binder that also binds HSA risks off-target effects. Tested across all designs to flag potential specificity issues early.
- Expression Yield: How much protein does each design produce? Higher yields are important for manufacturability. Measured for all designs.
What's included
- Binding affinities in triplicates (proxy KD derived from standardized 1:1 fitting)
- Expression and loading success rates
- Raw SPR sensorgrams & fits
- Neutralization assay results for top binders
- Designer method notes where provided
- Coming soon: HSA off-target binding data for top binders
- Expression Yield (ug/mL)
A note on fitting
Because NiV-G is a tetramer, most binding curves don't follow clean 1:1 kinetics. We observed bivalent and conformational binding effects throughout. Rather than force every curve into a model that doesn't fit, we first fit bivalent and conformational models to capture what's actually happening, then derived a standardized 1:1 proxy for fair ranking. All fits are included here so you can inspect the raw data, understand our methodology, or reanalyze with your own approach.