Proteins (2)
KPKaren Paco
id: hollow-shark-ash
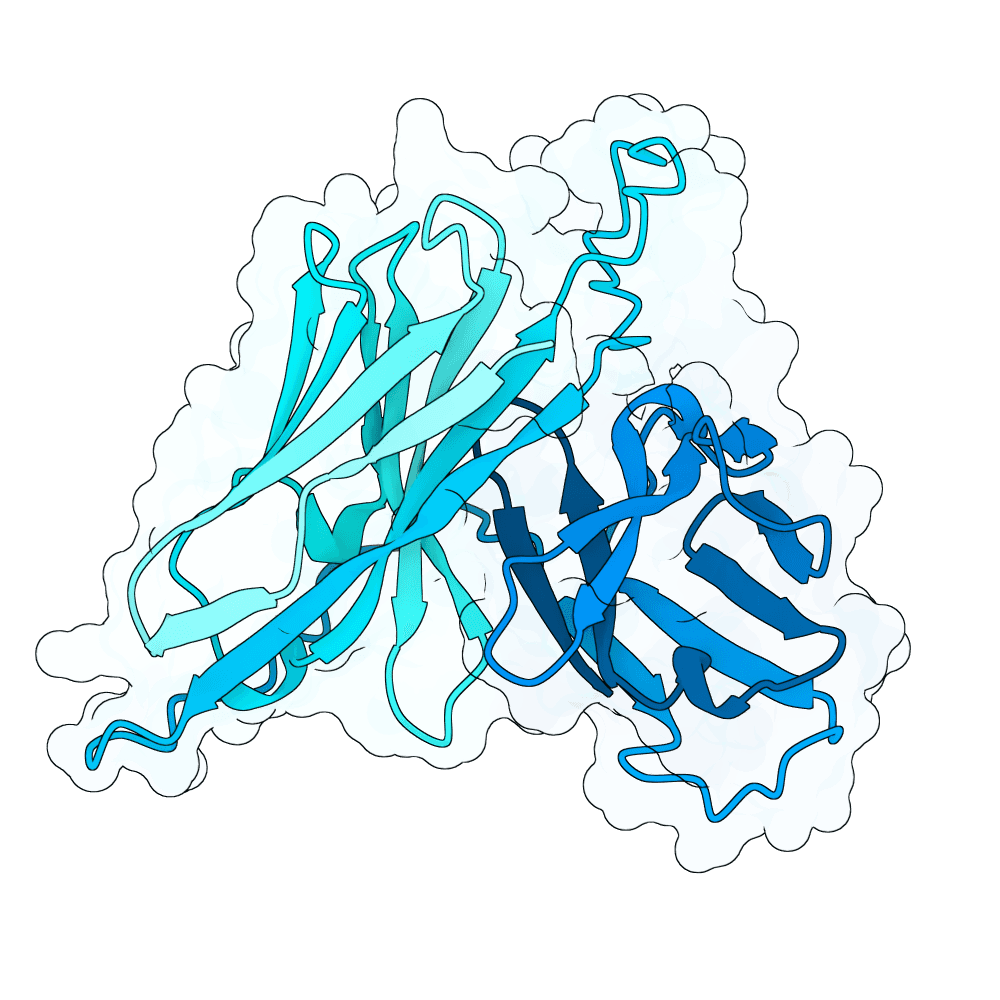
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
69.51
Expressed
True
Mol. Weight
26.4 kDa
AA Length
250
KPKaren Paco
id: deep-raven-topaz
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
69.37
Expressed
True
Mol. Weight
26.3 kDa
AA Length
249