Proteins (20)
id: hollow-jaguar-bronze
Nipah Virus Glycoprotein G
None
86.54
True
20.3 kDa
182
id: strong-vole-snow
Nipah Virus Glycoprotein G
None
79.59
False
20.4 kDa
182
id: hollow-quail-frost
Nipah Virus Glycoprotein G
None
73.34
True
19.9 kDa
182
id: mellow-raven-ivy
Nipah Virus Glycoprotein G
None
85.51
True
20.1 kDa
182
id: rough-ibis-orchid
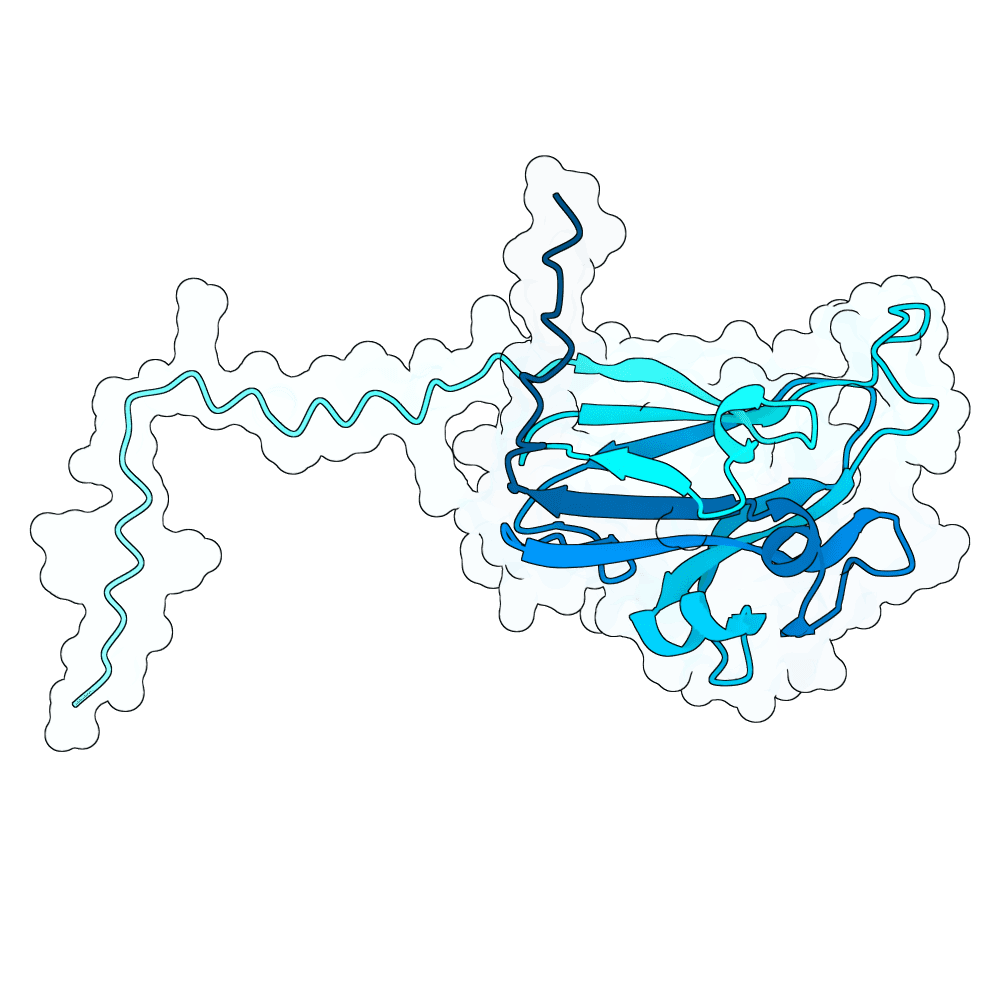
Nipah Virus Glycoprotein G
None
76.60
False
20.2 kDa
182
id: quiet-lion-clay
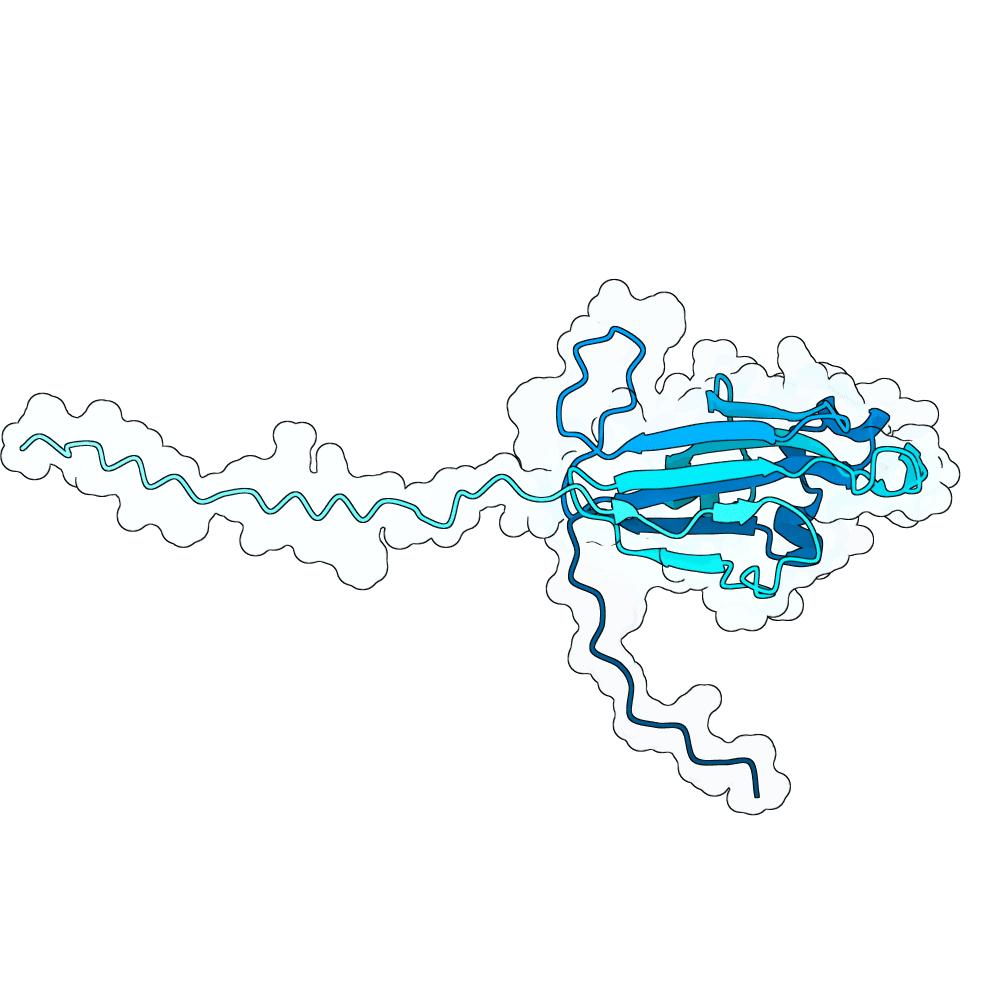
Nipah Virus Glycoprotein G
None
83.24
False
20.2 kDa
182
id: crimson-vole-birch
Nipah Virus Glycoprotein G
None
80.22
True
20.0 kDa
182
id: quiet-mole-onyx
Nipah Virus Glycoprotein G
None
78.81
True
19.9 kDa
182
id: vast-bat-ember
Nipah Virus Glycoprotein G
0.65
82.90
--
20.0 kDa
182
id: gentle-zebra-marble
Nipah Virus Glycoprotein G
0.77
72.88
--
20.0 kDa
182
id: steady-bee-reed
Nipah Virus Glycoprotein G
0.75
77.86
--
20.2 kDa
182
id: green-orca-vine
Nipah Virus Glycoprotein G
0.78
76.79
--
20.0 kDa
182
id: silent-tiger-opal
Nipah Virus Glycoprotein G
0.70
67.80
--
20.4 kDa
182
id: frozen-boar-pearl
Nipah Virus Glycoprotein G
0.74
79.55
--
20.2 kDa
182
id: ivory-bat-iron
Nipah Virus Glycoprotein G
0.81
75.30
--
20.3 kDa
182
id: radiant-kiwi-stone
Nipah Virus Glycoprotein G
0.78
71.05
--
20.3 kDa
182
id: solid-bear-granite
Nipah Virus Glycoprotein G
0.77
69.52
--
20.2 kDa
182
id: green-crane-pine
Nipah Virus Glycoprotein G
0.77
69.57
--
20.2 kDa
182
id: hollow-bear-dust
Nipah Virus Glycoprotein G
0.78
80.26
--
20.0 kDa
182
id: quick-panda-wave

Nipah Virus Glycoprotein G
0.75
67.03
--
20.0 kDa
182