Proteins (10)
id: vast-shark-crystal
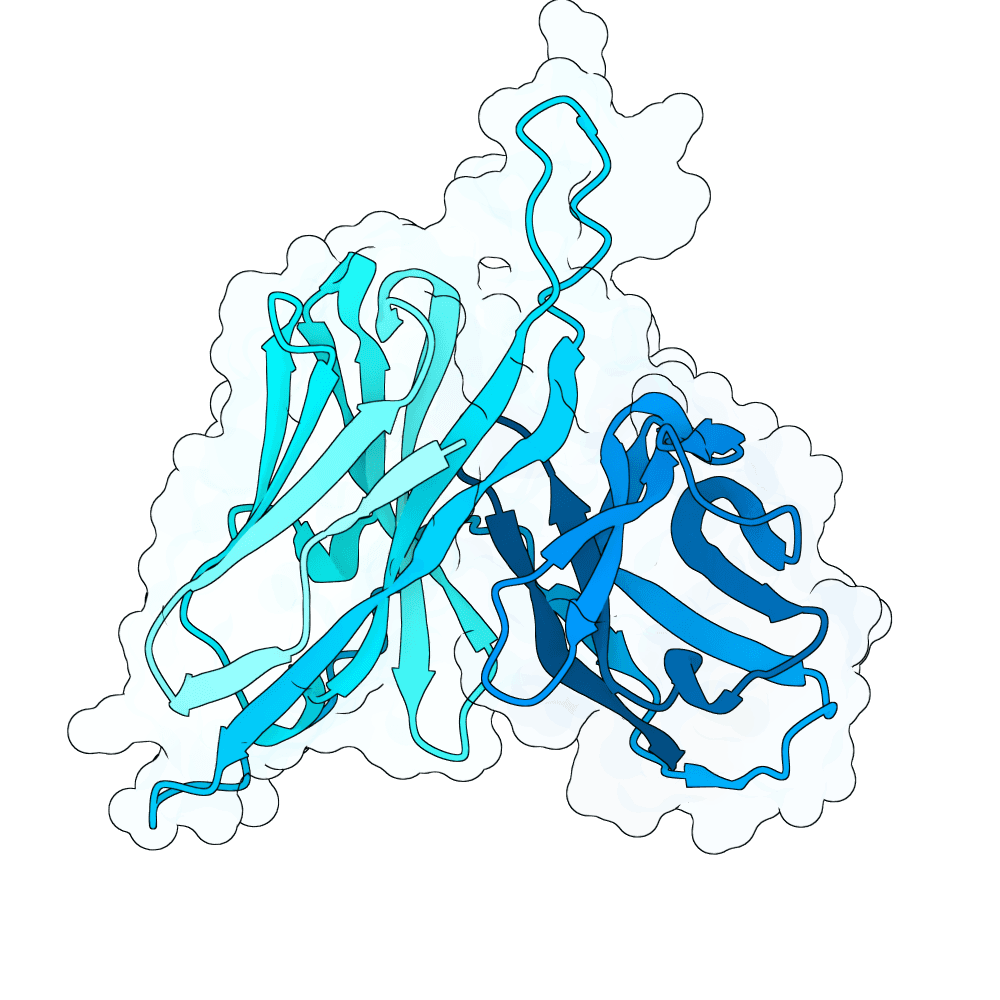
Nipah Virus Glycoprotein G
0.34
73.22
--
26.5 kDa
249
id: steady-deer-maple
Nipah Virus Glycoprotein G
0.03
73.68
--
26.5 kDa
249
id: quick-bison-dust
Nipah Virus Glycoprotein G
0.01
75.61
--
25.9 kDa
249
id: soft-jaguar-sand
Nipah Virus Glycoprotein G
0.22
76.85
--
25.8 kDa
249
id: strong-vole-willow
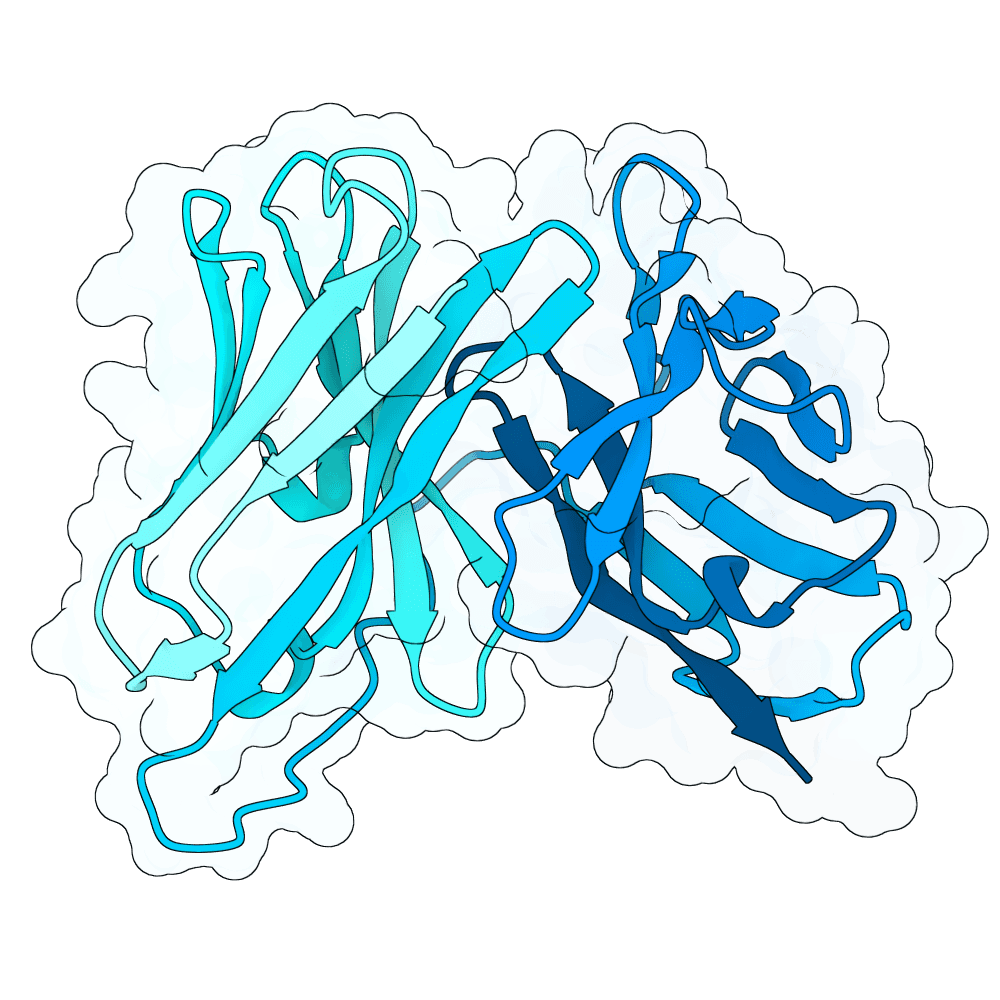
Nipah Virus Glycoprotein G
0.01
84.89
--
25.6 kDa
242
id: shy-panda-opal
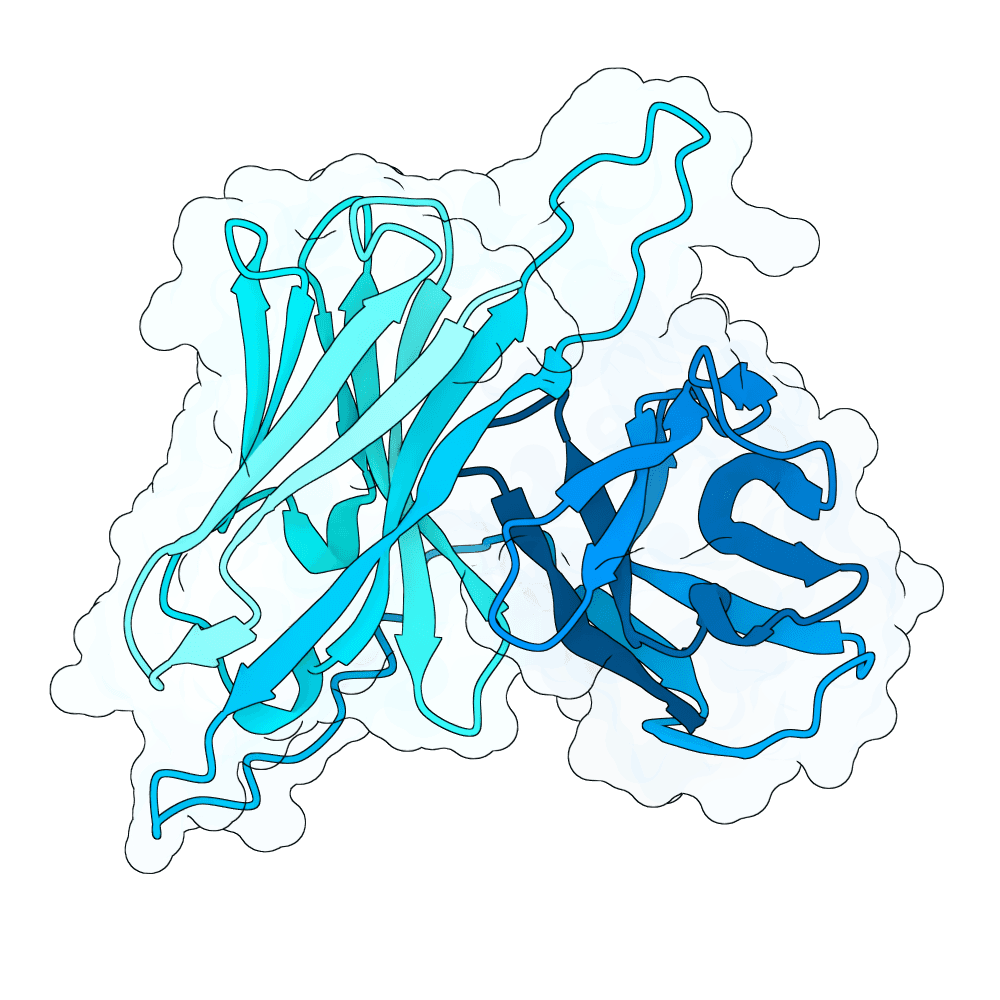
Nipah Virus Glycoprotein G
0.00
77.88
--
26.2 kDa
248
id: dark-panda-flint
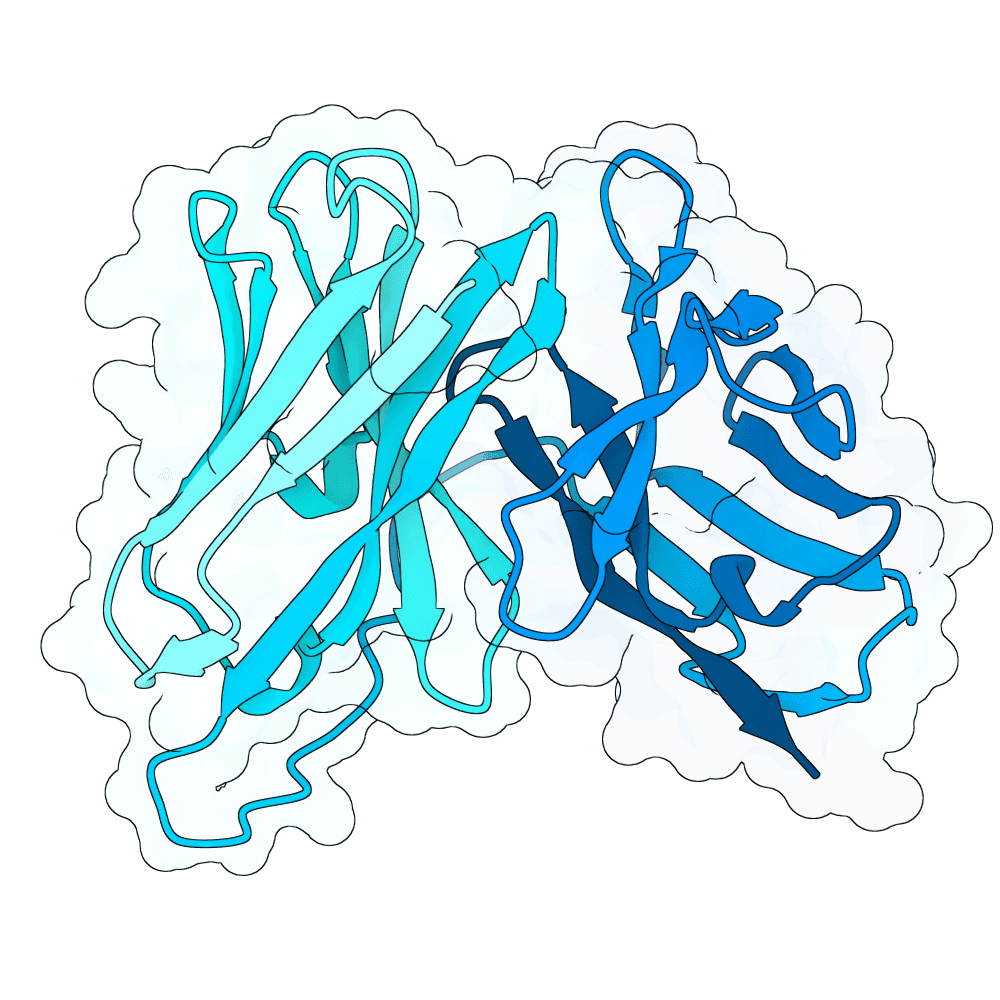
Nipah Virus Glycoprotein G
0.00
83.61
--
25.9 kDa
242
id: crimson-kiwi-ember
Nipah Virus Glycoprotein G
0.00
74.95
--
25.9 kDa
250
id: dark-swan-vine
Nipah Virus Glycoprotein G
0.00
81.26
--
26.2 kDa
248
id: hollow-dove-quartz
Nipah Virus Glycoprotein G
0.44
77.42
--
25.9 kDa
250