Proteins (10)
id: crimson-crow-bronze
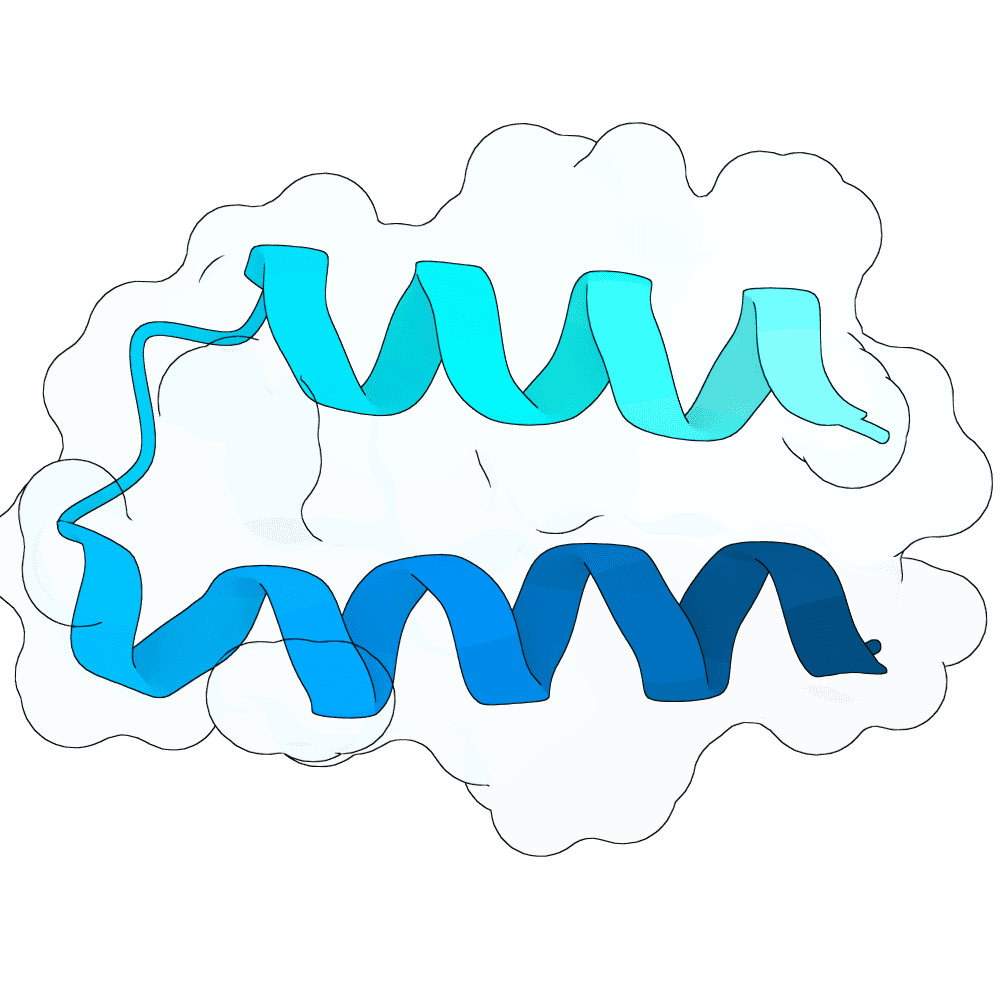
Nipah Virus Glycoprotein G
None
74.57
True
3.6 kDa
35
id: azure-bear-lava
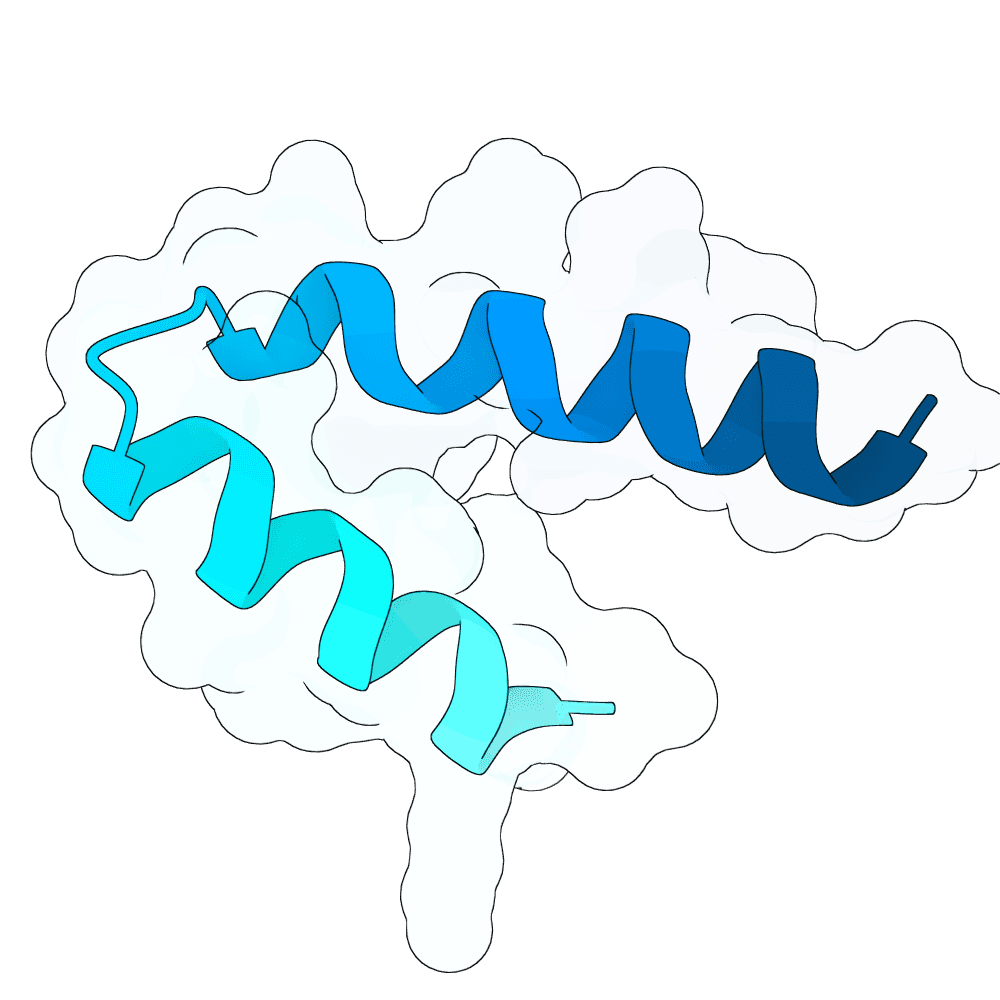
Nipah Virus Glycoprotein G
None
59.64
True
3.6 kDa
35
id: crimson-hawk-ice
Nipah Virus Glycoprotein G
None
57.99
True
3.7 kDa
35
id: lunar-fox-opal
Nipah Virus Glycoprotein G
None
62.06
True
3.5 kDa
35
id: amber-falcon-pearl
Nipah Virus Glycoprotein G
None
59.59
True
3.6 kDa
35
id: gentle-crane-snow
Nipah Virus Glycoprotein G
None
59.43
True
3.8 kDa
35
id: gentle-hawk-wave
Nipah Virus Glycoprotein G
None
59.94
True
3.8 kDa
35
id: strong-eagle-snow
Nipah Virus Glycoprotein G
None
60.11
True
3.6 kDa
35
id: quiet-otter-bronze
Nipah Virus Glycoprotein G
None
59.85
True
3.6 kDa
35
id: gentle-cobra-ruby
Nipah Virus Glycoprotein G
0.87
53.59
--
4.0 kDa
35