Proteins (8)
id: soft-seal-frost
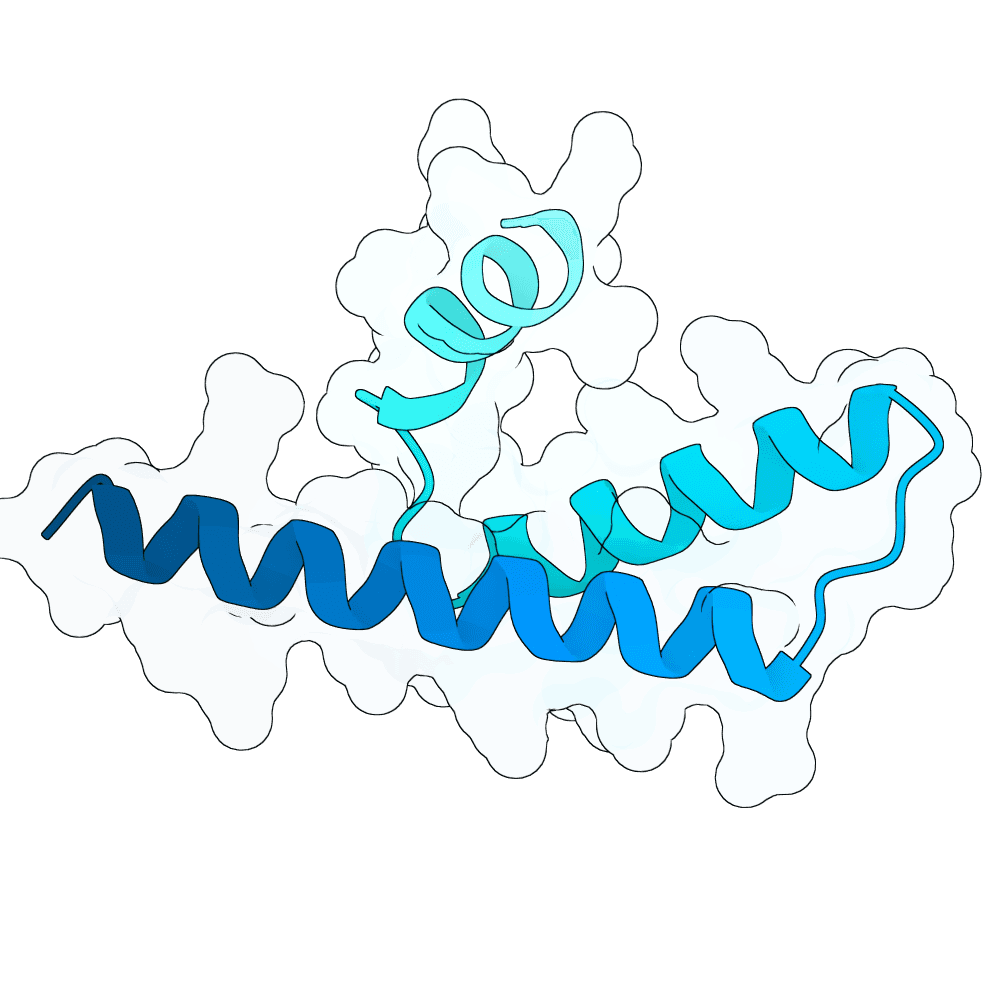
Nipah Virus Glycoprotein G
None
60.60
True
7.0 kDa
64
id: golden-seal-thorn
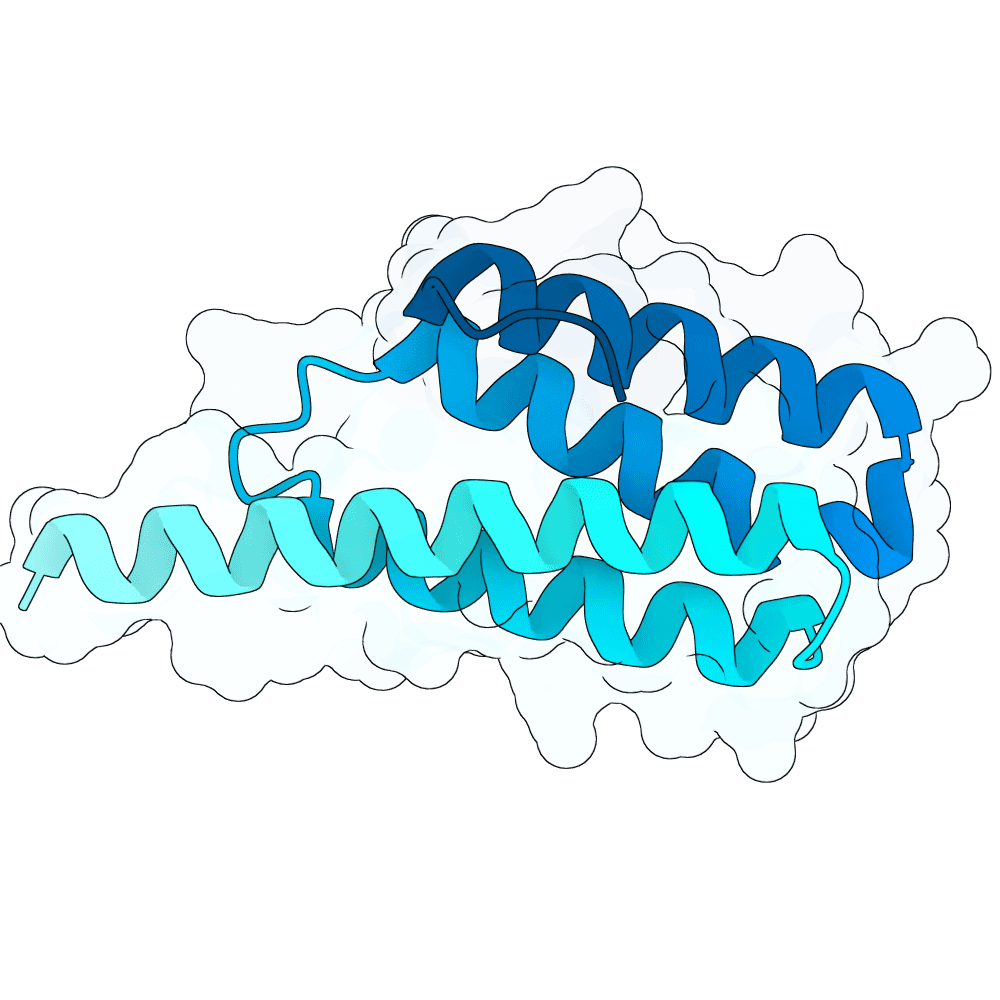
Nipah Virus Glycoprotein G
None
87.75
True
10.6 kDa
101
id: pale-wolf-oak
Nipah Virus Glycoprotein G
None
83.58
True
6.9 kDa
64
id: vast-bee-sand
Nipah Virus Glycoprotein G
None
85.37
True
7.1 kDa
64
id: quick-lion-pearl
Nipah Virus Glycoprotein G
None
86.48
True
10.9 kDa
101
id: crimson-heron-snow
Nipah Virus Glycoprotein G
None
86.91
True
10.8 kDa
101
id: lunar-toad-bronze
Nipah Virus Glycoprotein G
0.71
86.78
--
11.0 kDa
101
id: rough-bison-ivy
Nipah Virus Glycoprotein G
0.87
86.94
--
10.7 kDa
101