Proteins (39)
id: steady-bat-crystal

PD-L1
Weak
6.1e-6 M
True
9.3 kDa
80
id: ivory-mole-wave

PD-L1
Weak
4.9e-6 M
True
9.2 kDa
80
id: misty-cat-stone

Multiple (2)
Weak (2), Strong (2)
True
9.5 kDa
80
id: brisk-tiger-oak

Multiple (2)
Weak (2), Strong (2)
True
9.2 kDa
80
id: quick-falcon-stone

Multiple (2)
Weak (2), Strong (2)
True
9.3 kDa
80
id: small-goat-ice

Multiple (2)
Weak (2), Strong (2)
True
9.3 kDa
80
id: bright-swan-lotus

IL-7Ra
Weak
1.8e-6 M
True
9.7 kDa
80
id: crimson-tiger-flint

PD-L1
Weak
1.4e-6 M
True
9.2 kDa
80
id: scarlet-kiwi-thorn

Multiple (2)
Weak (2), Strong (2)
True
9.3 kDa
80
id: bright-otter-reed

Multiple (2)
Weak (2), Strong (2)
True
9.3 kDa
80
id: solid-ram-wave

Multiple (2)
Weak (2), Strong (2)
True
9.4 kDa
80
id: golden-heron-quartz

PD-L1
Medium
5.2e-7 M
True
9.0 kDa
80
id: gentle-raven-marble

IL-7Ra
Medium
4.2e-7 M
True
9.4 kDa
80
id: golden-cat-lava

Multiple (2)
Medium (2), Strong (2)
True
9.3 kDa
80
id: young-bear-cypress

IL-7Ra
Medium
3.1e-7 M
True
9.1 kDa
80
id: noble-moth-lava

PD-L1
Medium
2.5e-7 M
True
9.5 kDa
80
id: jade-ram-plume

PD-L1
Medium
1.1e-7 M
True
9.4 kDa
80
id: crimson-mole-quartz

PD-L1
Medium
1.1e-7 M
True
9.1 kDa
80
id: steady-eagle-oak
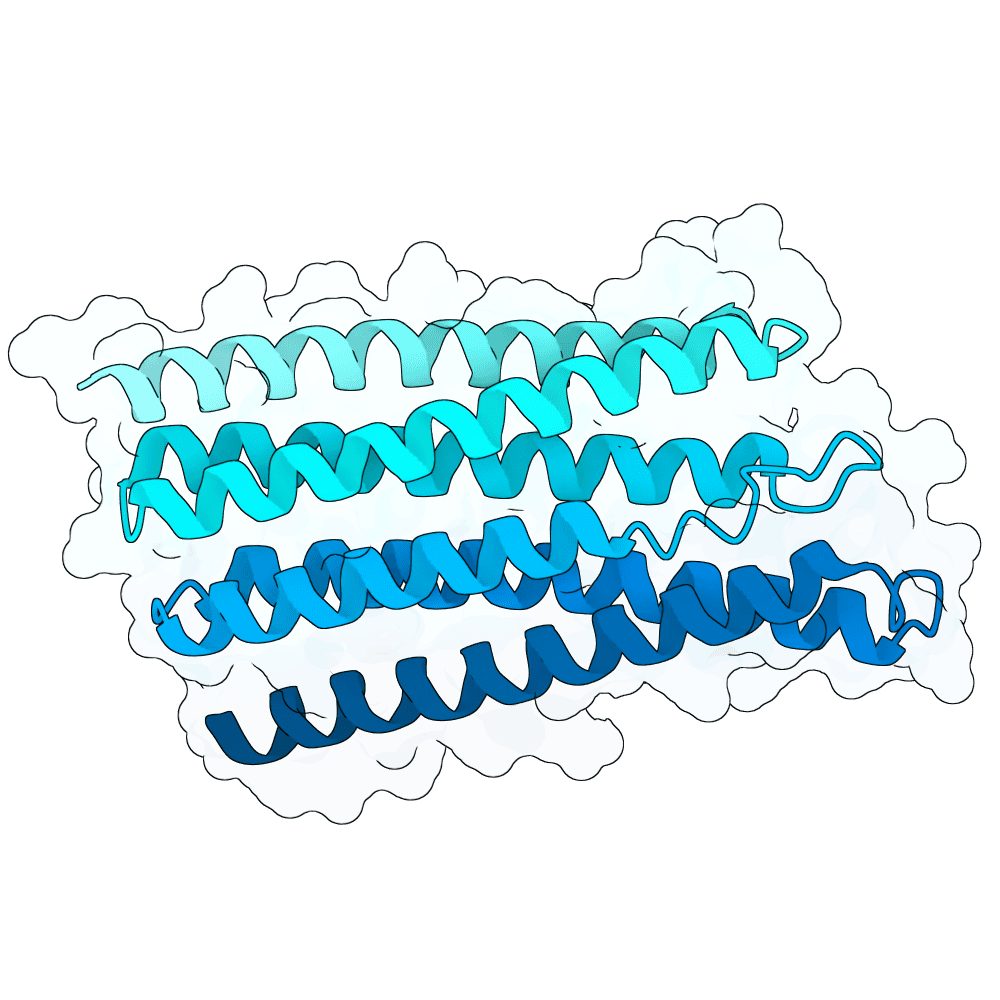
Multiple (2)
Medium (6)
True
24.2 kDa
220
id: soft-goat-oak

IL-7Ra
Medium
7.1e-8 M
True
9.9 kDa
80
id: frozen-bear-flint
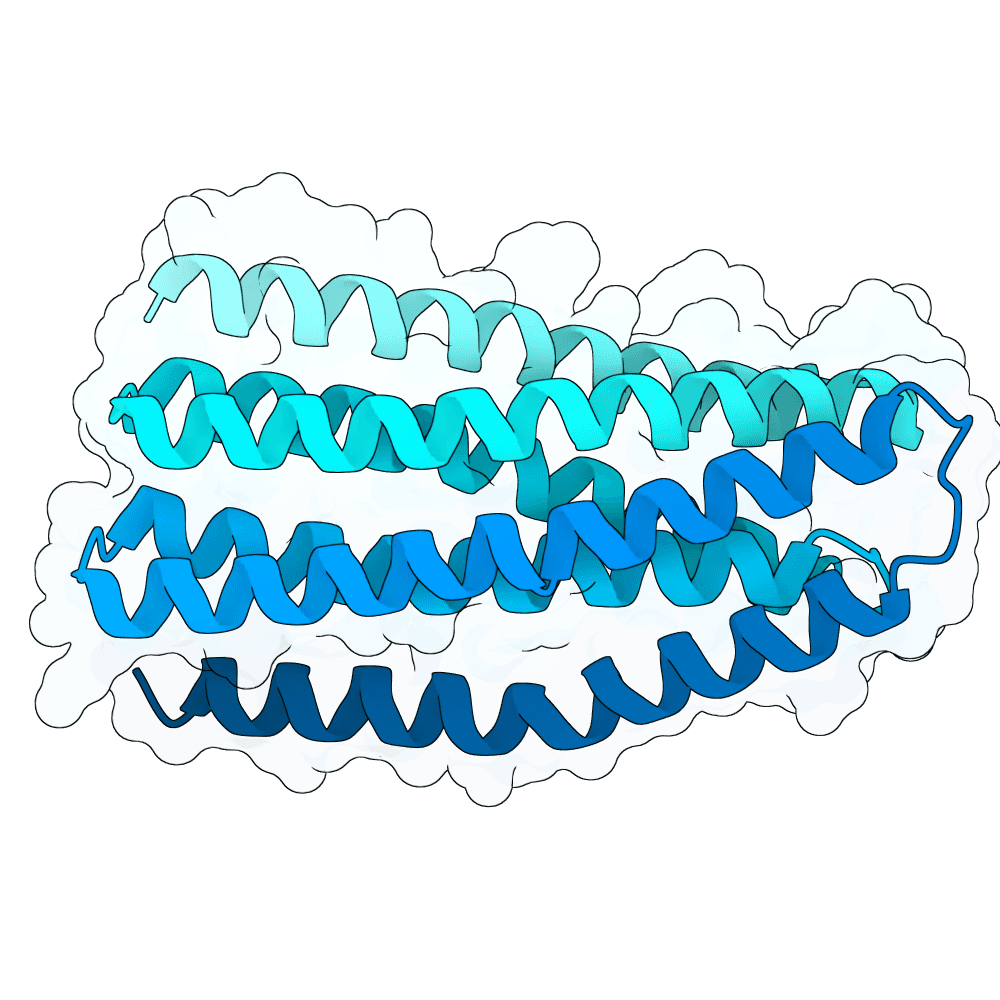
Multiple (2)
Strong (6)
True
23.1 kDa
220