Proteins (10)
id: pale-vole-vine
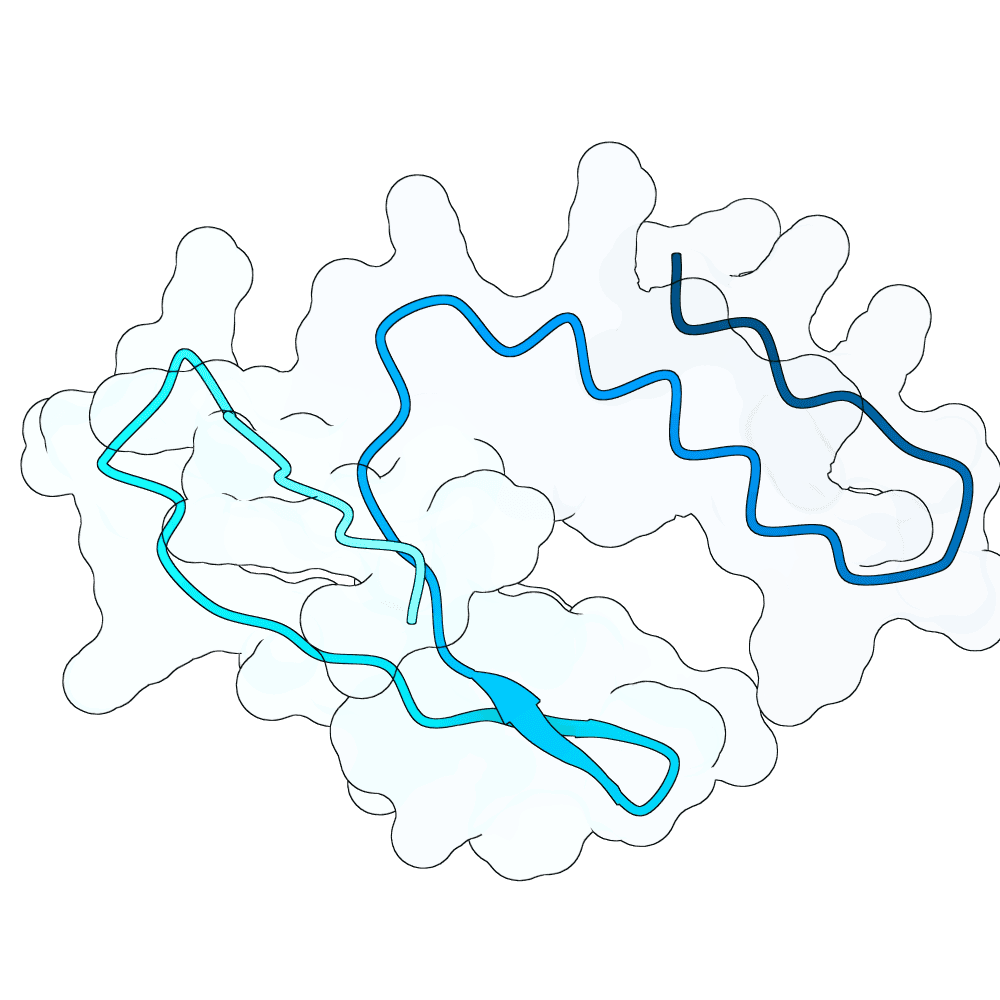
Nipah Virus Glycoprotein G
None
33.57
False
6.7 kDa
52
id: soft-shark-clay
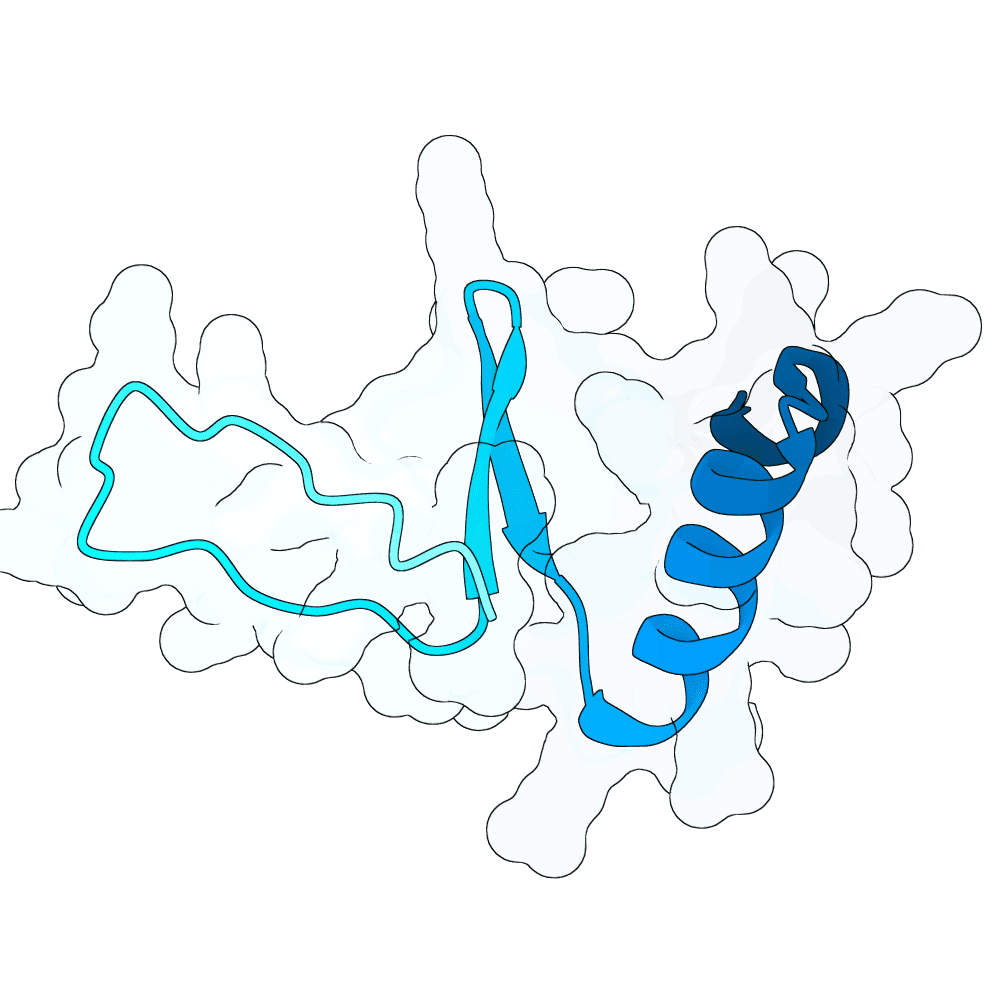
Nipah Virus Glycoprotein G
None
44.77
True
6.1 kDa
51
id: ivory-goat-sand
Nipah Virus Glycoprotein G
None
66.40
True
6.1 kDa
51
id: green-mole-clay
Nipah Virus Glycoprotein G
None
41.09
True
6.5 kDa
51
id: amber-heron-pine
Nipah Virus Glycoprotein G
None
51.55
False
6.4 kDa
51
id: noble-jaguar-ash
Nipah Virus Glycoprotein G
None
36.79
True
6.4 kDa
51
id: soft-fox-ruby
Nipah Virus Glycoprotein G
None
49.12
True
6.4 kDa
52
id: mellow-otter-leaf
Nipah Virus Glycoprotein G
None
48.30
True
6.2 kDa
51
id: gentle-moth-lotus
Nipah Virus Glycoprotein G
0.41
44.93
--
6.5 kDa
51
id: swift-seal-reed
Nipah Virus Glycoprotein G
0.00
51.21
--
6.1 kDa
51