Proteins (10)
id: azure-mole-vine
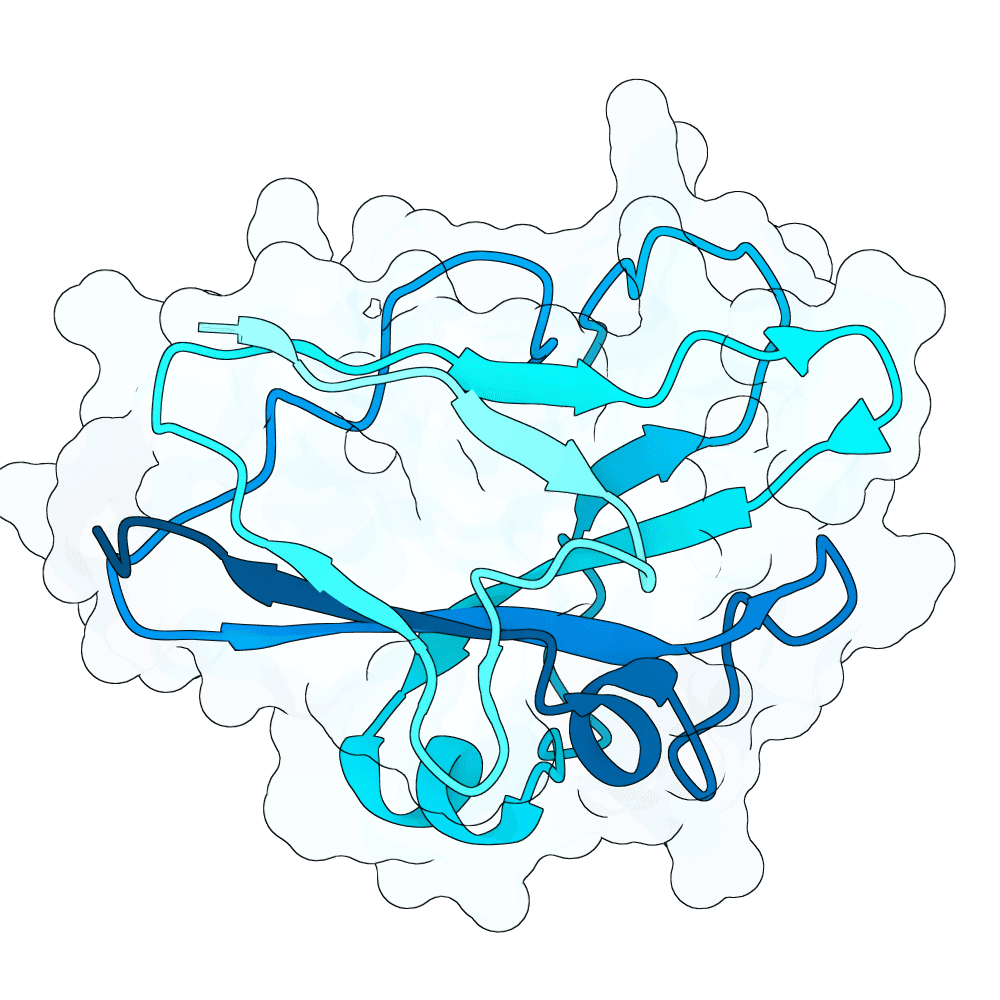
Nipah Virus Glycoprotein G
None
58.87
True
15.2 kDa
137
id: pale-vole-quartz
Nipah Virus Glycoprotein G
None
35.66
False
15.5 kDa
137
id: calm-bee-dust
Nipah Virus Glycoprotein G
None
64.60
True
15.1 kDa
137
id: solid-boar-ivy
Nipah Virus Glycoprotein G
None
23.28
False
15.5 kDa
137
id: jade-kiwi-thorn
Nipah Virus Glycoprotein G
0.01
83.44
--
15.3 kDa
137
id: deep-lynx-iron
Nipah Virus Glycoprotein G
0.09
54.97
--
15.2 kDa
137
id: pale-seal-iron
Nipah Virus Glycoprotein G
0.02
62.90
--
15.1 kDa
137
id: dark-quail-pine
Nipah Virus Glycoprotein G
0.79
79.96
--
15.0 kDa
137
id: scarlet-ox-pearl
Nipah Virus Glycoprotein G
0.31
80.17
--
15.1 kDa
137
id: ivory-bison-pearl
Nipah Virus Glycoprotein G
0.34
64.75
--
15.1 kDa
137