Proteins (31)
id: golden-boar-flint

EGFR
Weak
2.2e-6 M
True
6.7 kDa
58
id: azure-crow-cypress

Nipah Virus Glycoprotein G
None
78.48
True
8.0 kDa
72
id: jade-deer-cedar
EGFR
None
80.75
True
6.4 kDa
58
id: brisk-yak-ruby
EGFR
None
83.95
True
6.4 kDa
58
id: quick-moth-plume
EGFR
None
80.76
True
6.4 kDa
58
id: silver-boar-jade
EGFR
None
83.70
True
6.4 kDa
58
id: solid-hawk-willow
EGFR
Weak
78.99
True
6.4 kDa
58
id: vast-quail-cypress
EGFR
None
82.19
True
6.4 kDa
58
id: bright-otter-iron
EGFR
None
83.27
True
5.8 kDa
52
id: violet-vole-opal
EGFR
None
86.54
True
5.4 kDa
49
id: noble-fox-maple
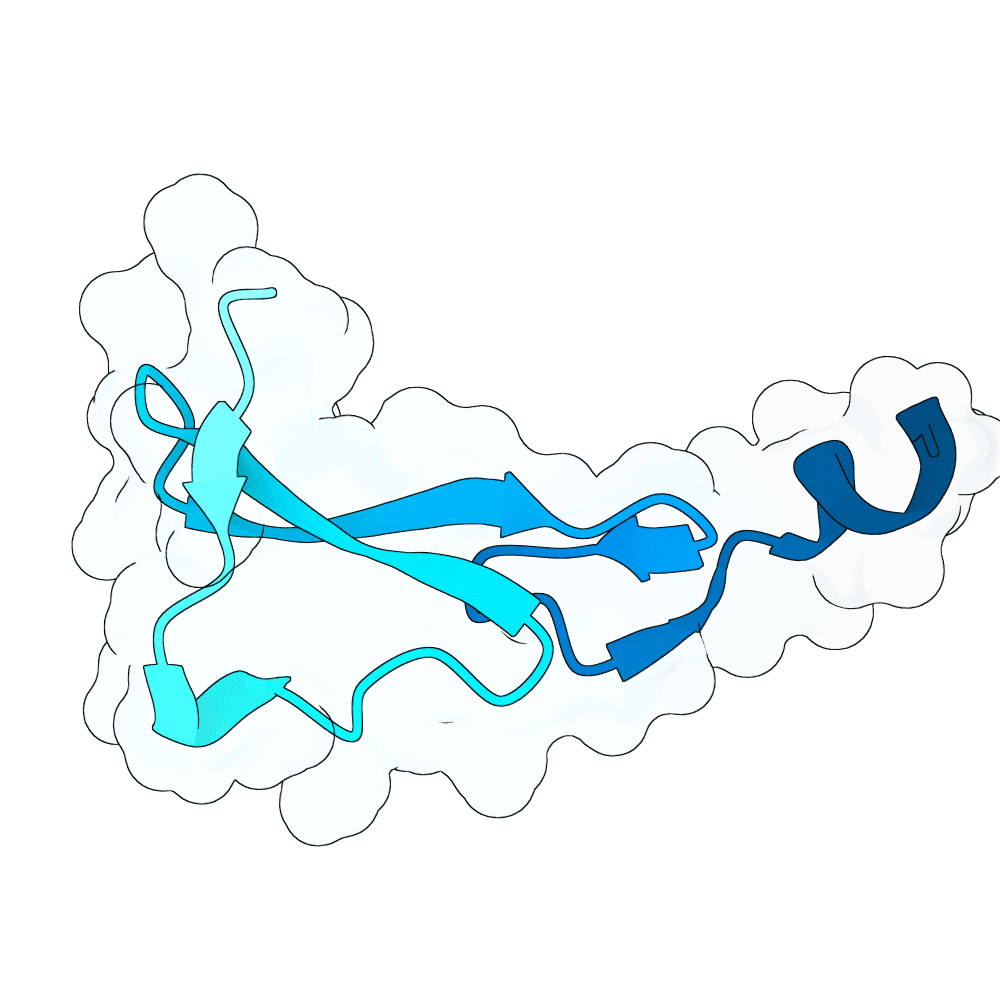
EGFR
None
80.88
True
5.8 kDa
52
id: rough-falcon-cypress

EGFR
None
81.18
True
6.5 kDa
58
id: quiet-orca-ruby
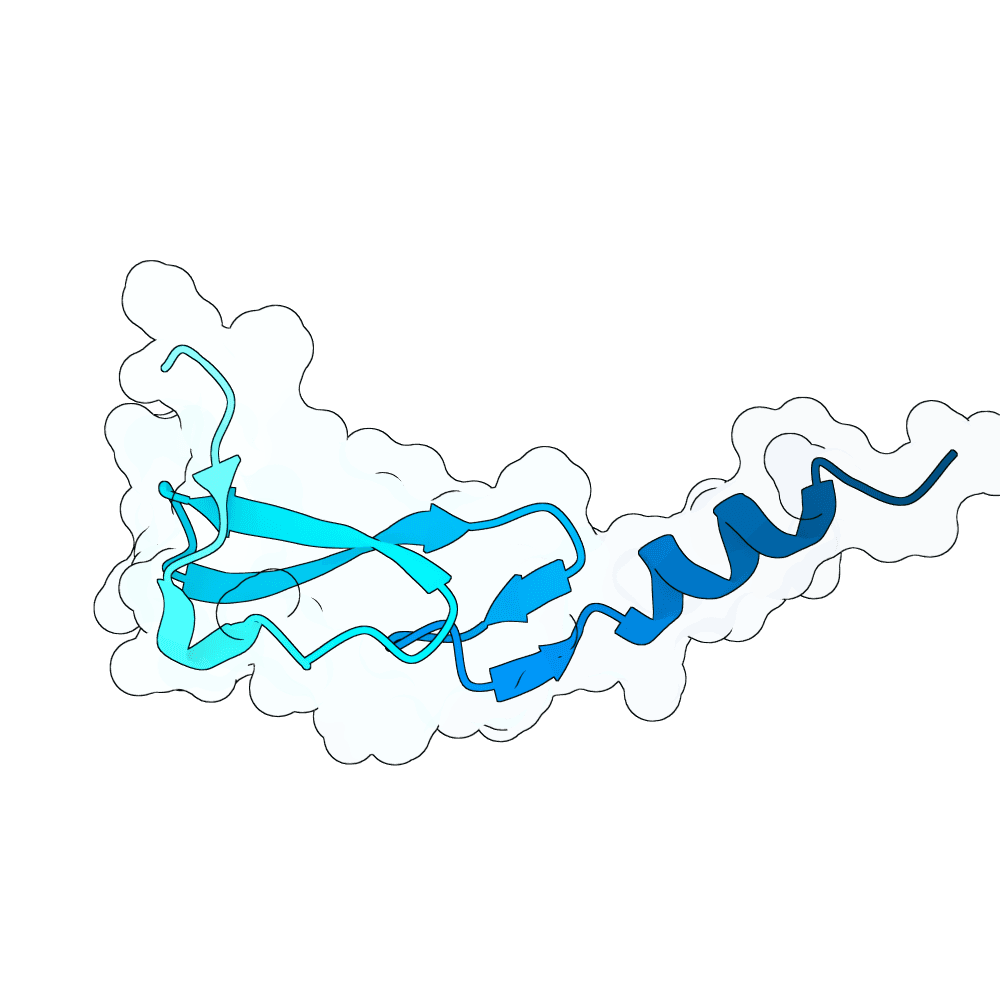
EGFR
None
81.16
True
6.6 kDa
58
id: calm-crane-bronze
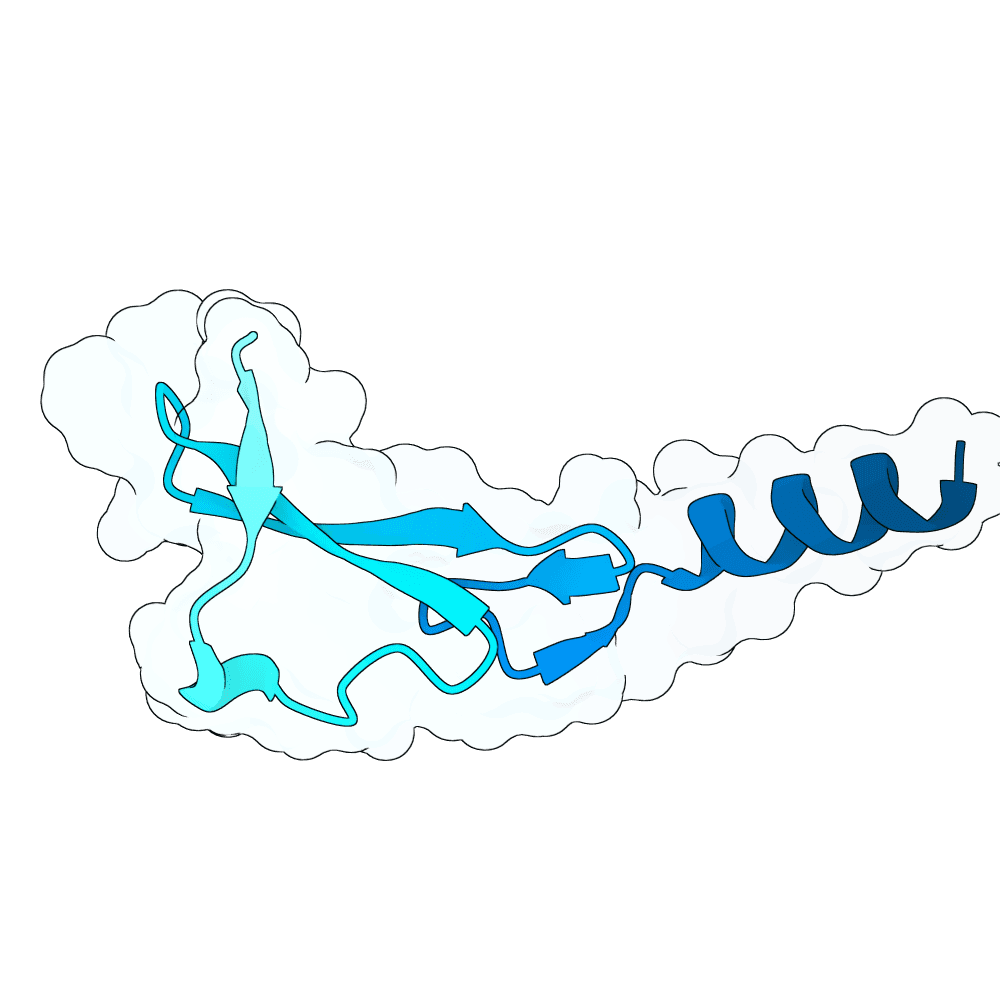
EGFR
None
86.73
True
6.1 kDa
58
id: steady-wolf-stone

EGFR
None
70.45
True
6.4 kDa
58
id: solid-gecko-cloud
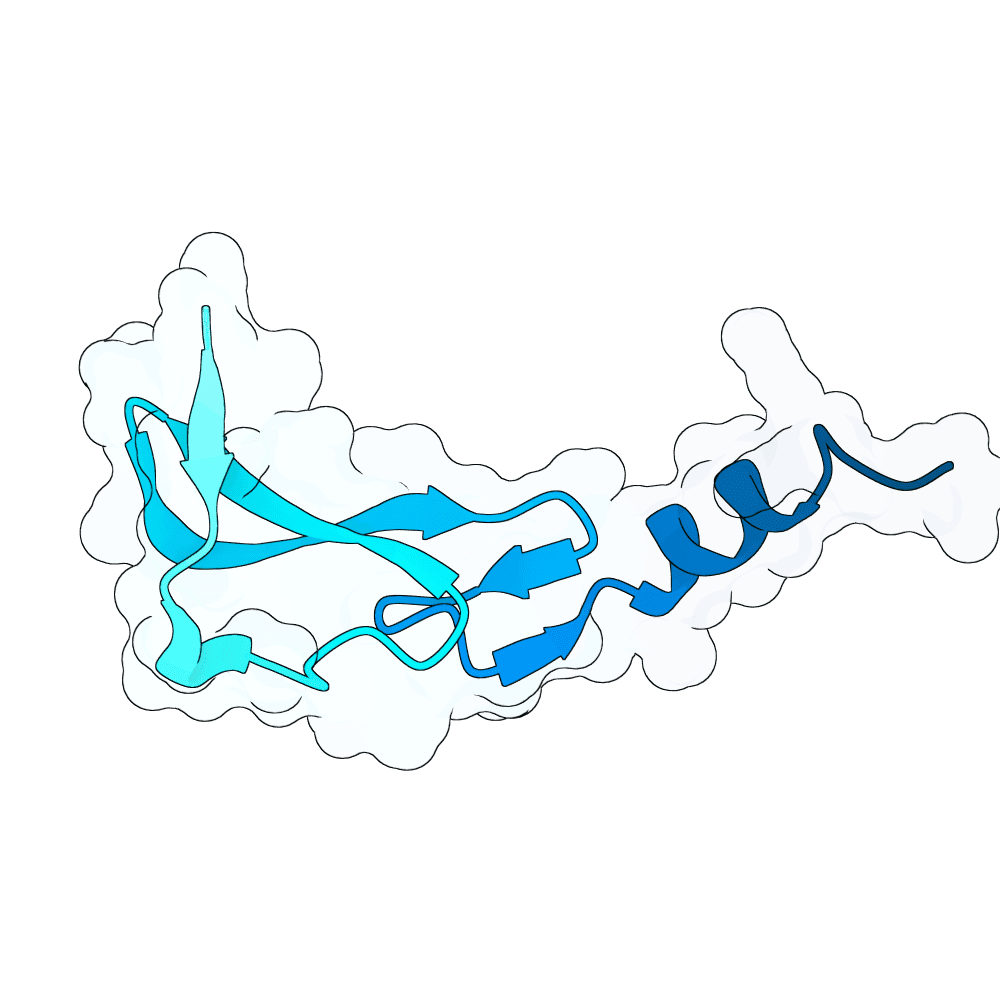
EGFR
None
77.53
True
6.4 kDa
58
id: swift-dove-opal
EGFR
None
77.89
True
6.4 kDa
58
id: quick-hawk-topaz
EGFR
None
80.28
True
6.5 kDa
58
id: brisk-swan-frost
EGFR
None
77.56
True
5.5 kDa
49
id: strong-panda-lava
EGFR
None
79.94
True
6.5 kDa
58
id: mellow-panther-ember
EGFR
None
76.89
True
6.6 kDa
58