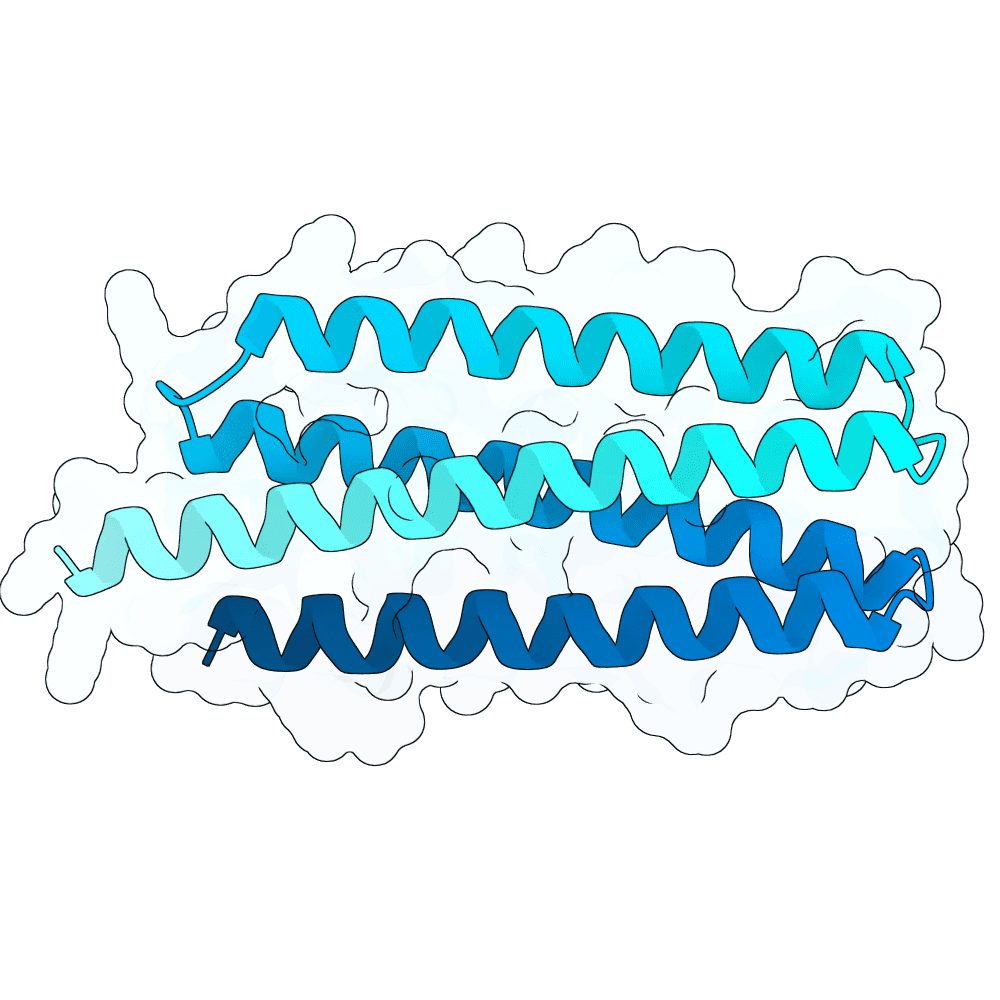
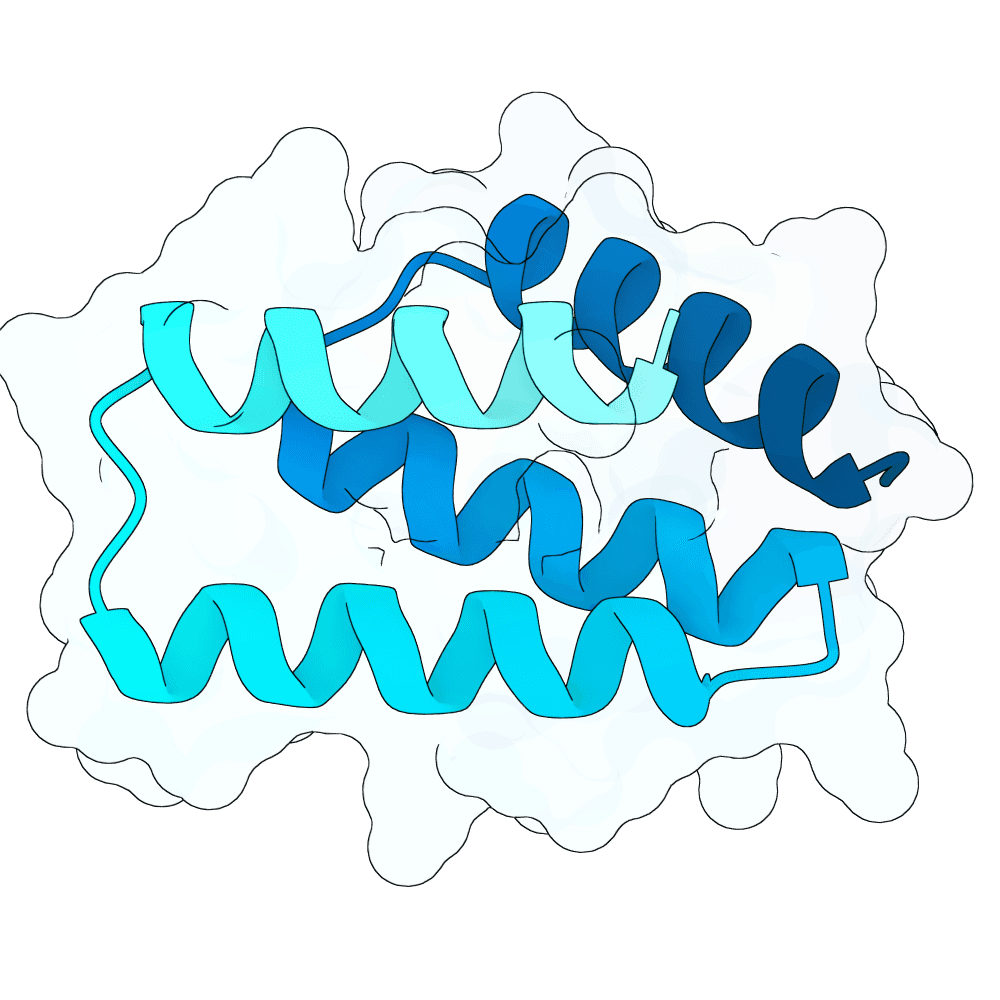
Description
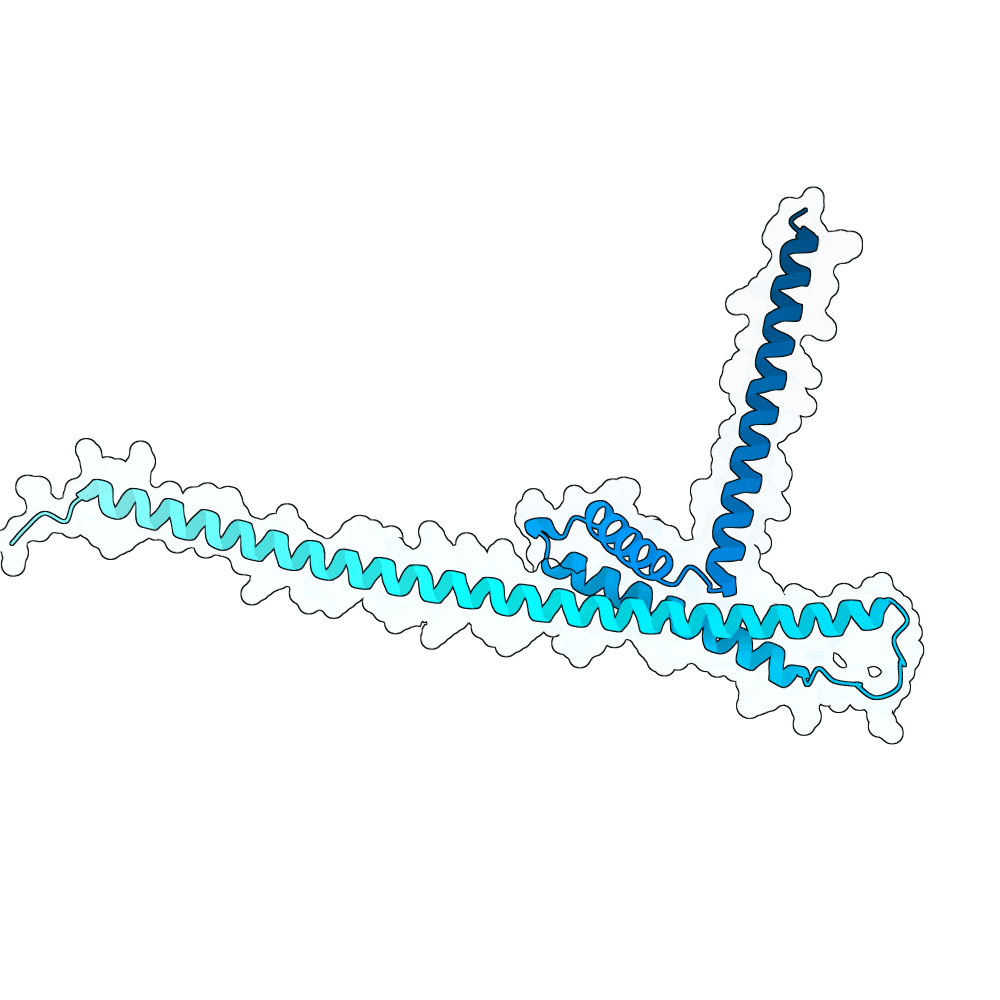
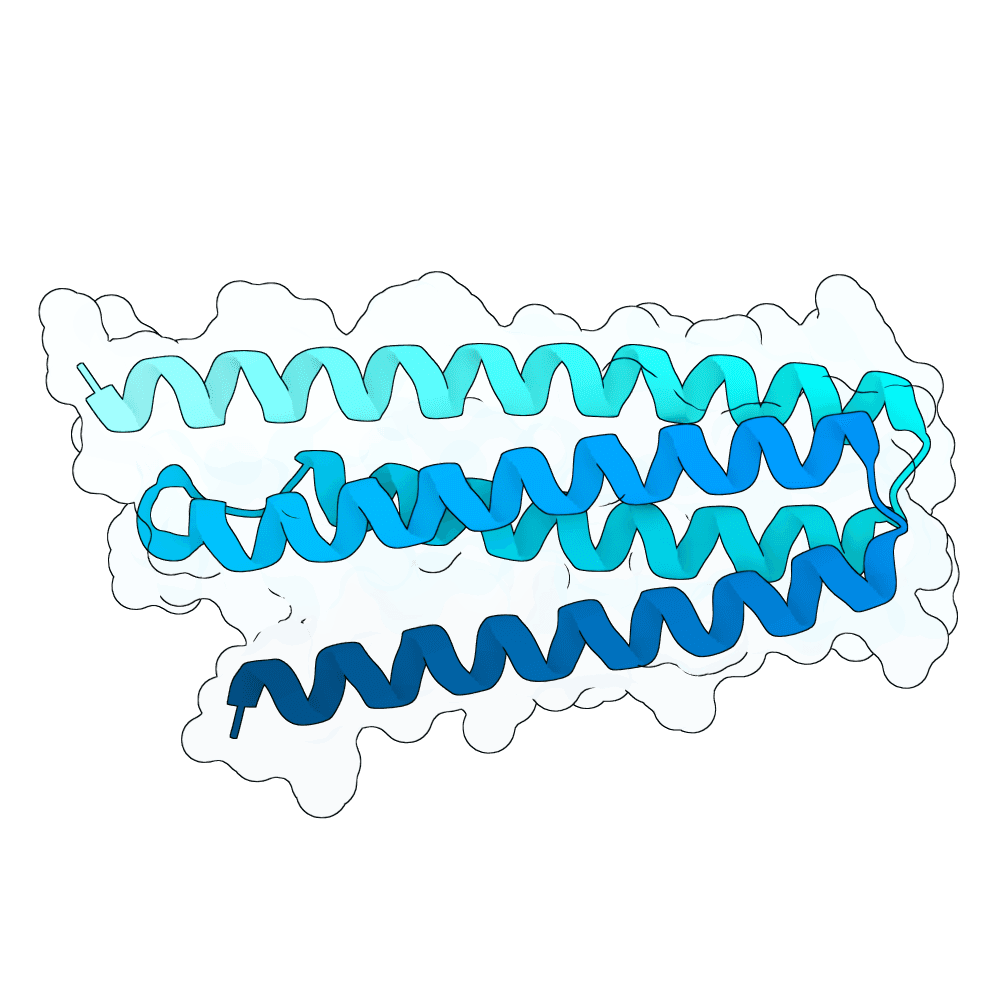
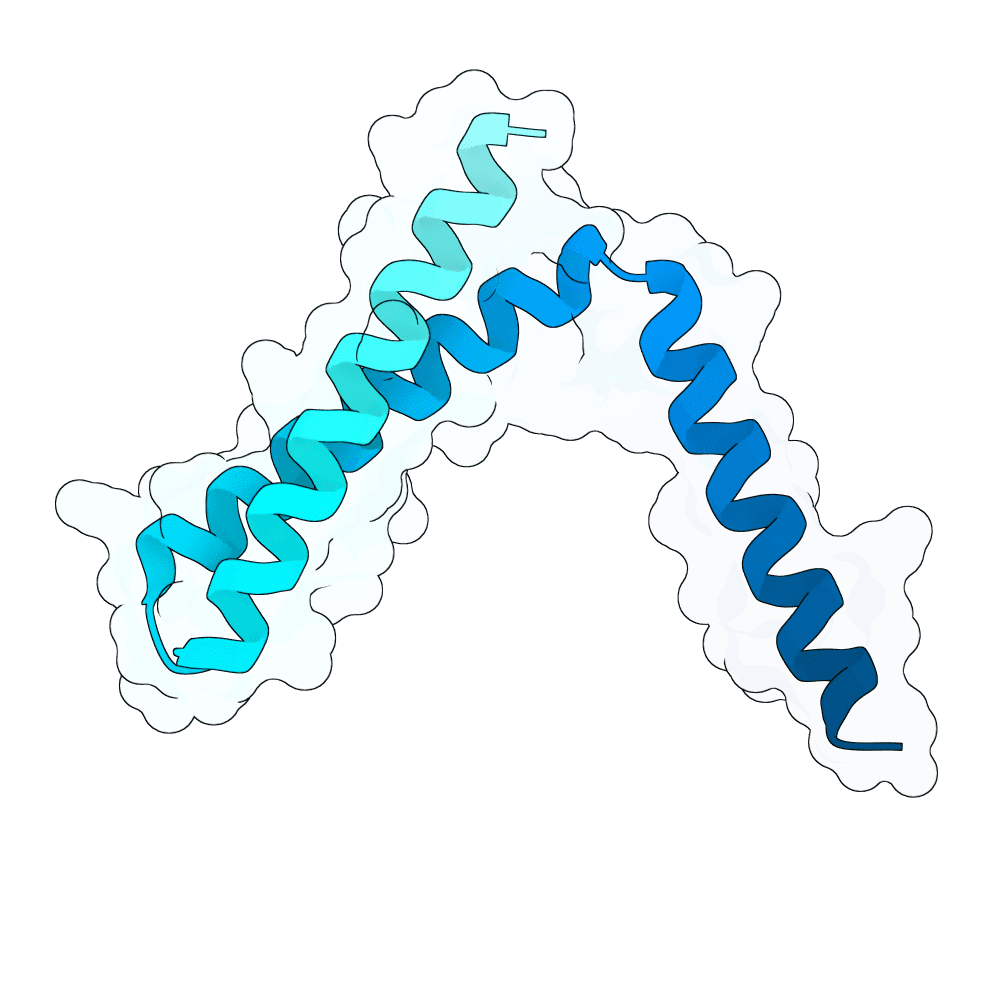
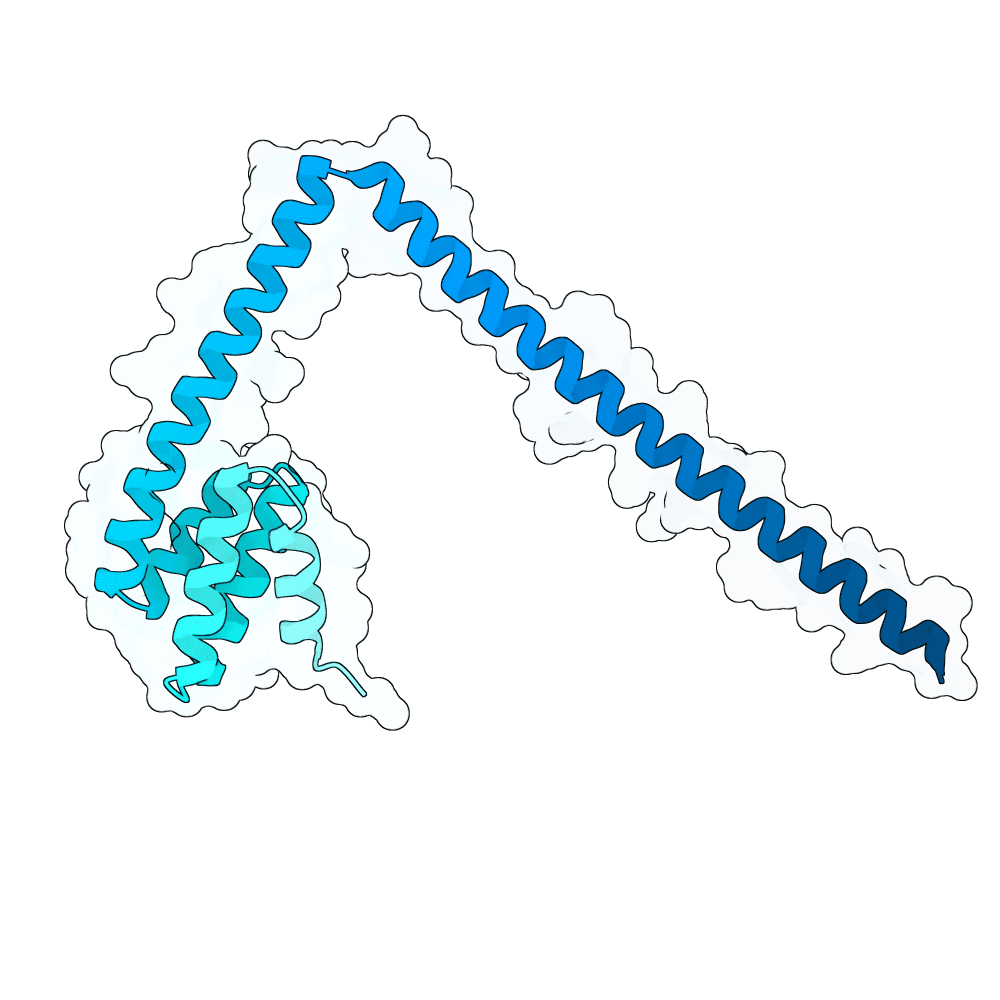
We first performed Molecular Dynamics (MD) Simulations on the Glycoprotein G (including its sugars; 2 microseconds-long) to get a wider variety of starting structures. We assumed that, by sampling different conformations, we could reveal metastable binding pockets not present in the crystal structure that could be targetable. After clustering the MD trajectories, we selected 25 centroid structures that represented the entire ensemble captured by the simulation as targets. We then ran RFdiffusion to generate a total of 5,000 binders against either the experimentally-determined target structure or an MD-derived structure. Hotspots in the target structures were determined by visual inspection and the use of the PeSTo software. Inverse folding was performed using CARBonAra with its default parameters. For each RfDiffusion-generated backbone, 5 sequences were designed. After this, AlphaFold3 was applied to fold the complexes. The ipSAE metric was used as a ranking criterion for the submission. The top 10 binder-target complexes were visually inspected, and all non-paired cysteines in the designs were replaced with either alanine or serine, depending on how exposed these residues were.
Target structures: 2VSM (https://www.rcsb.org/structure/2VSM)
MD engine: GROMACS 2024.02 (https://www.sciencedirect.com/science/article/pii/S2352711015000059)
Structure-binder generation: RFDiffusion (https://www.nature.com/articles/s41586-023-06415-8)
Inverse folding: CARBonAra (https://www.biorxiv.org/content/10.1101/2023.06.19.545381v1.full.pdf)
Validation: AlphaFold3 (https://www.nature.com/articles/s41586-024-07487-w)