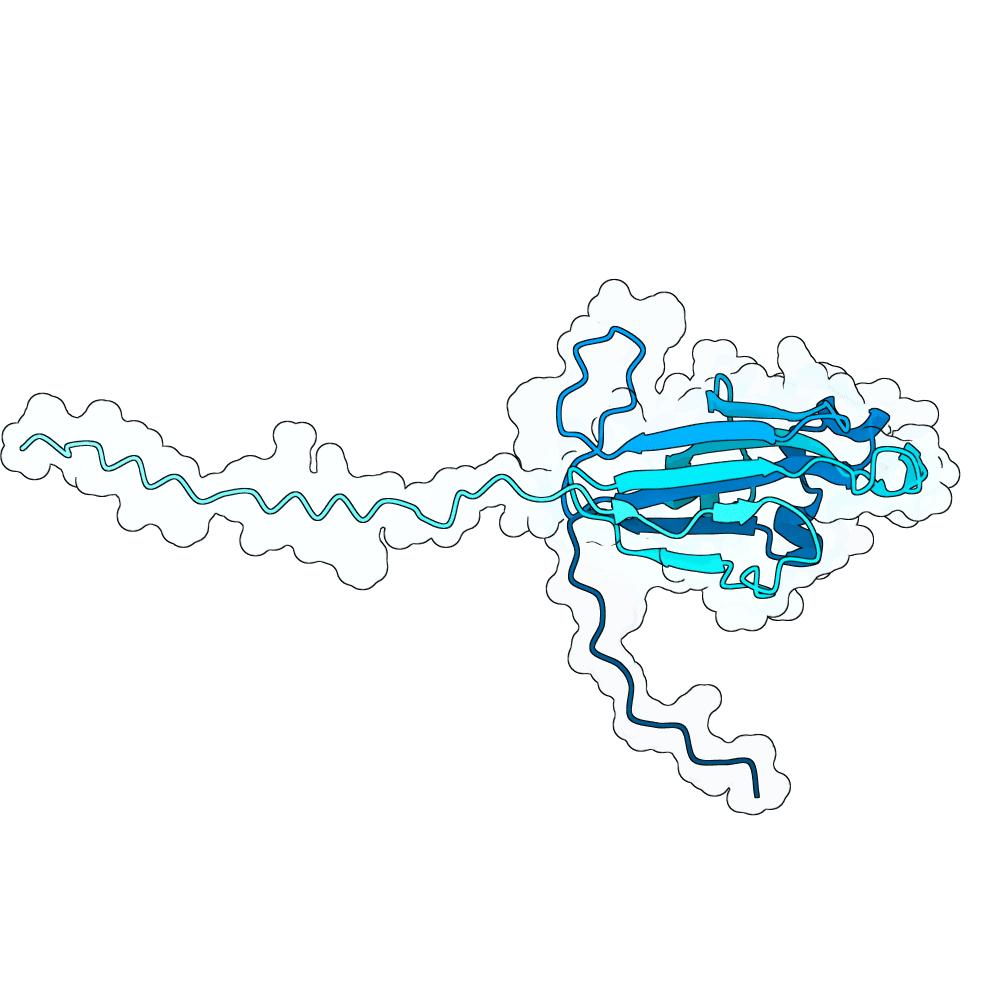
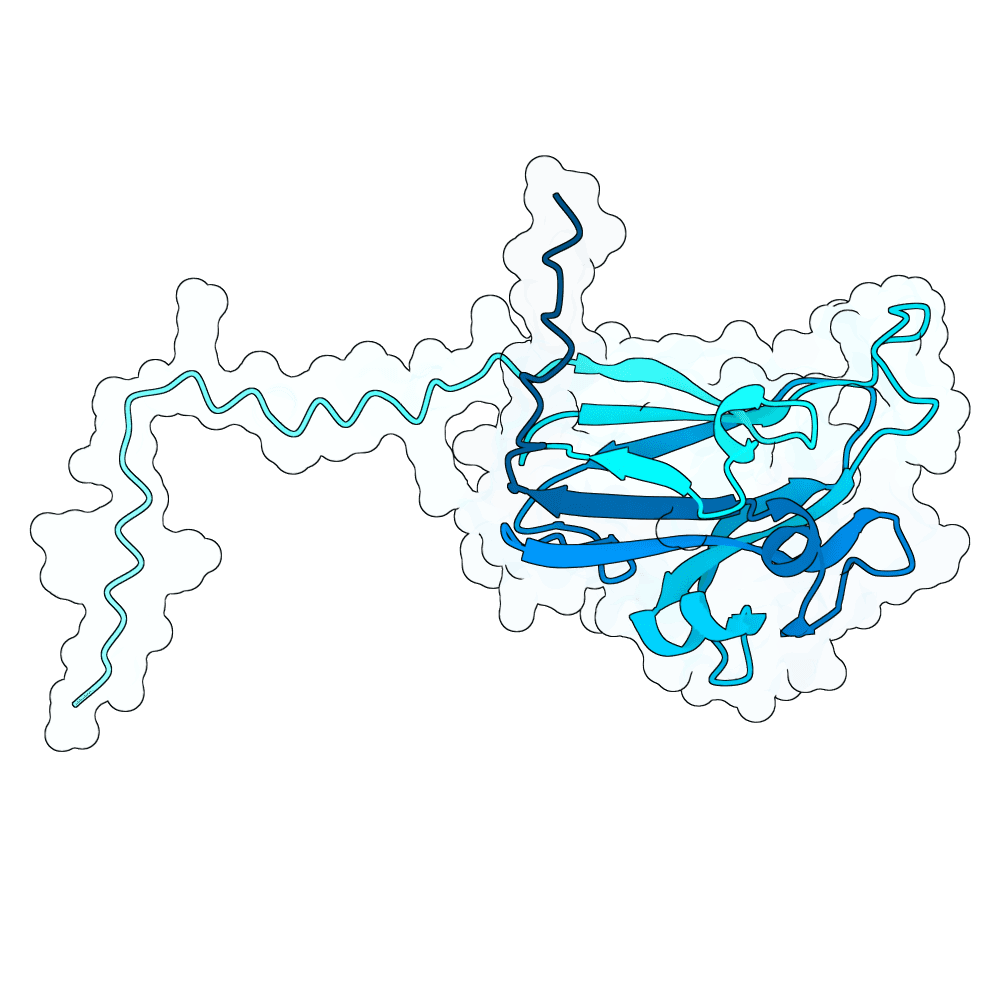
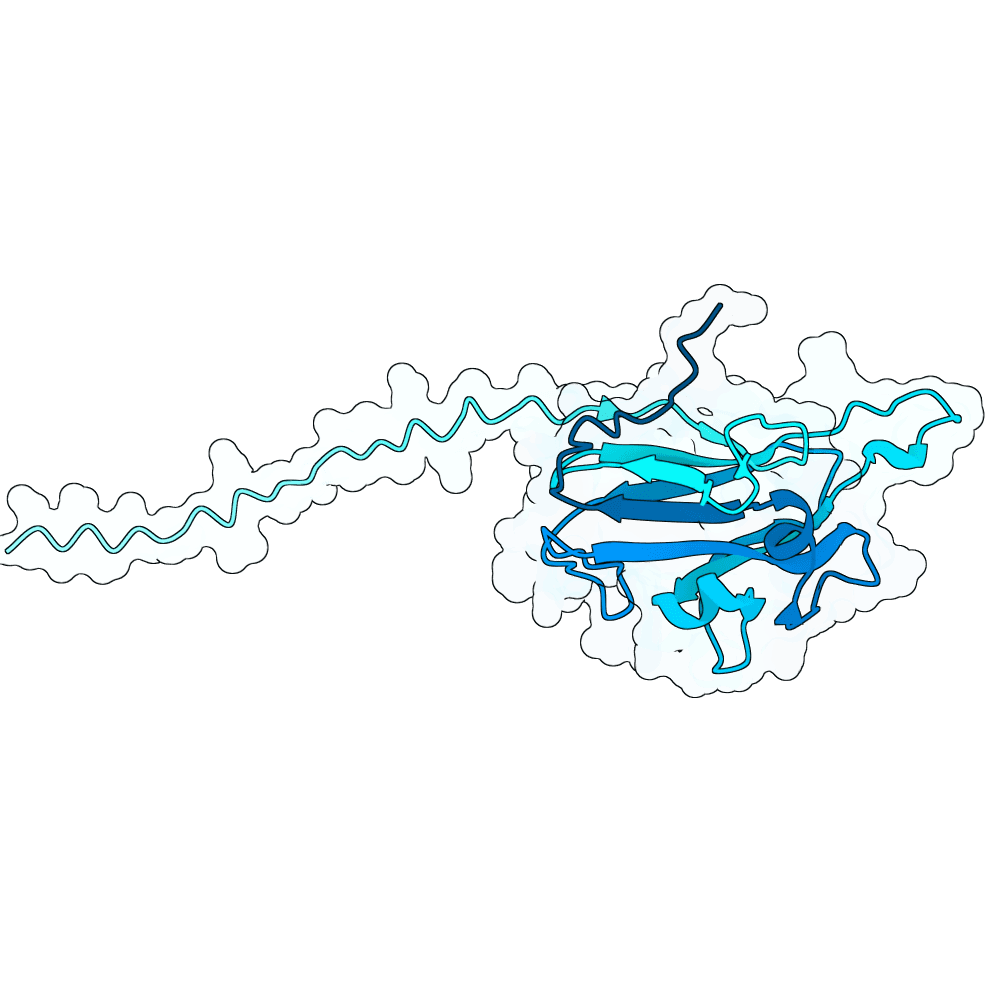
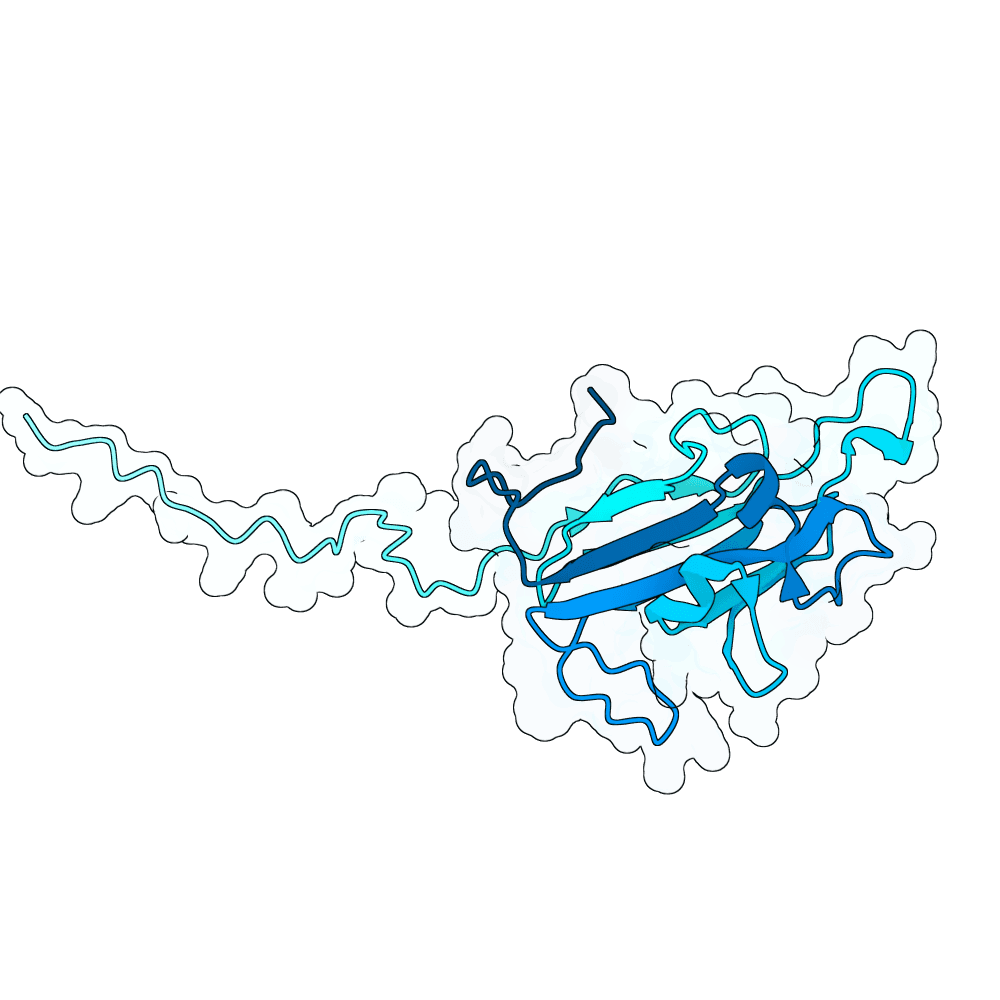
Description
The Synthyra Protein Design Environment (PDE) is an iterative, modular, and efficient pipeline that can effectively engineer proteins in a multi-objective fitness landscape using sequence-only protein modeling. Thus far, it has been used to design de novo and lead optimization protein binders.
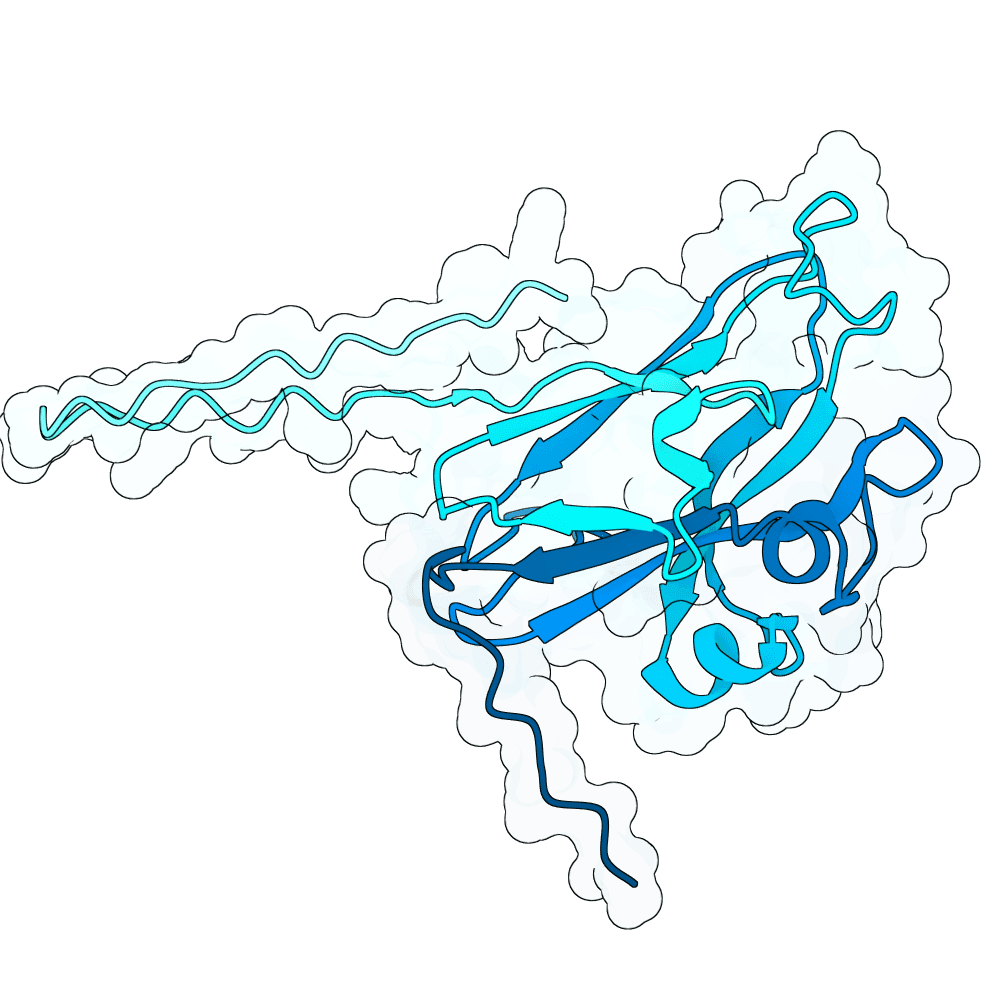
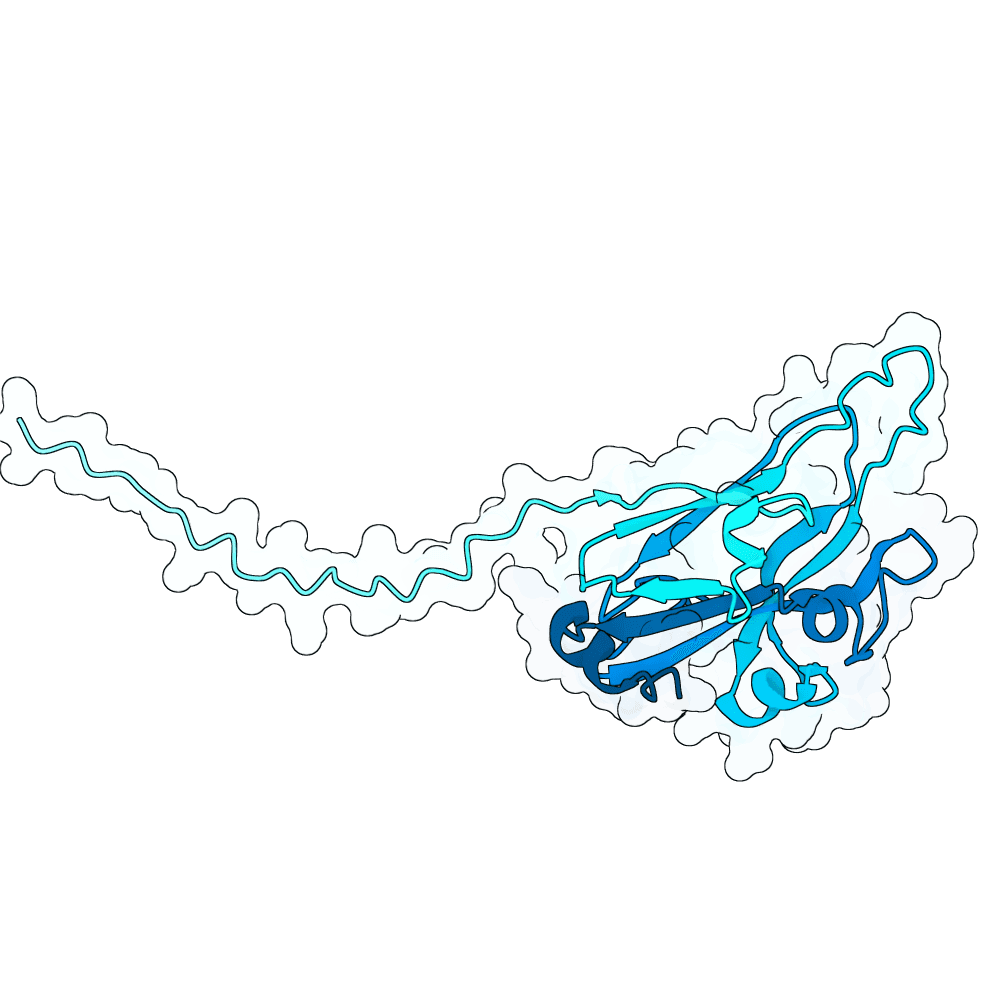
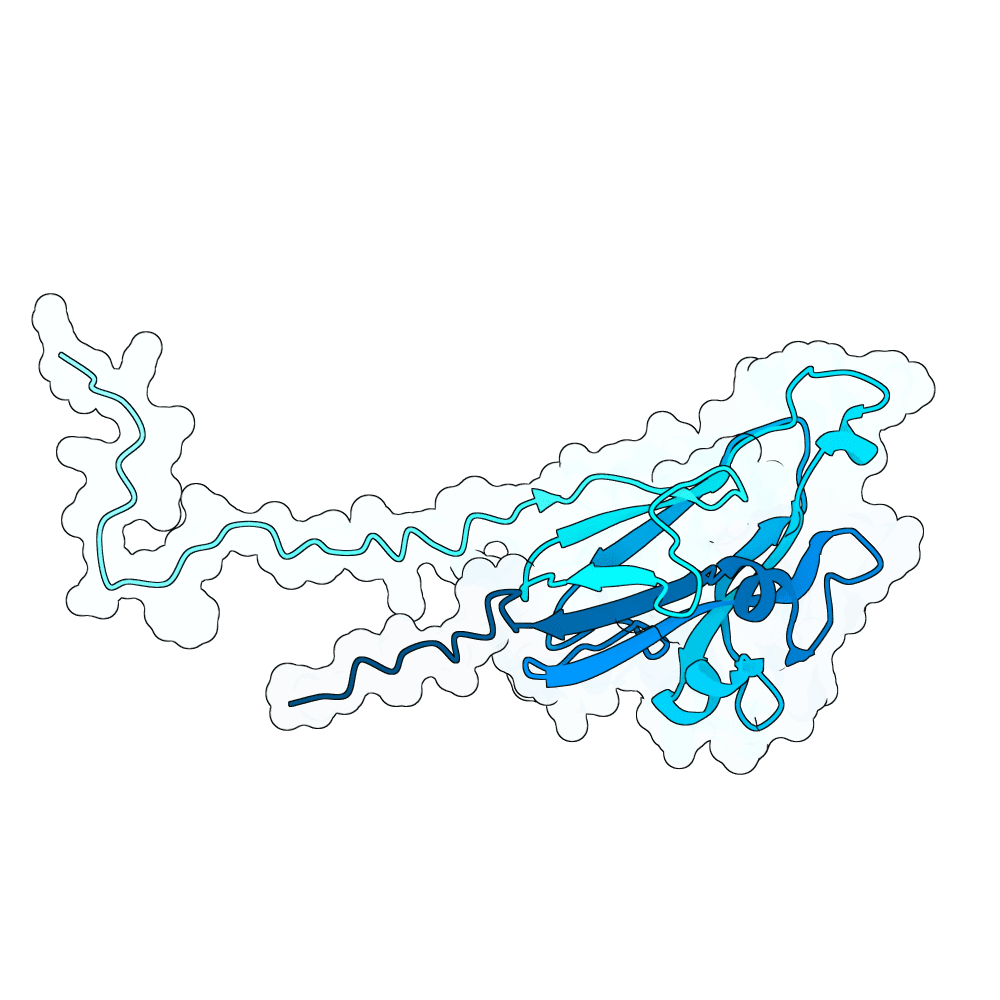
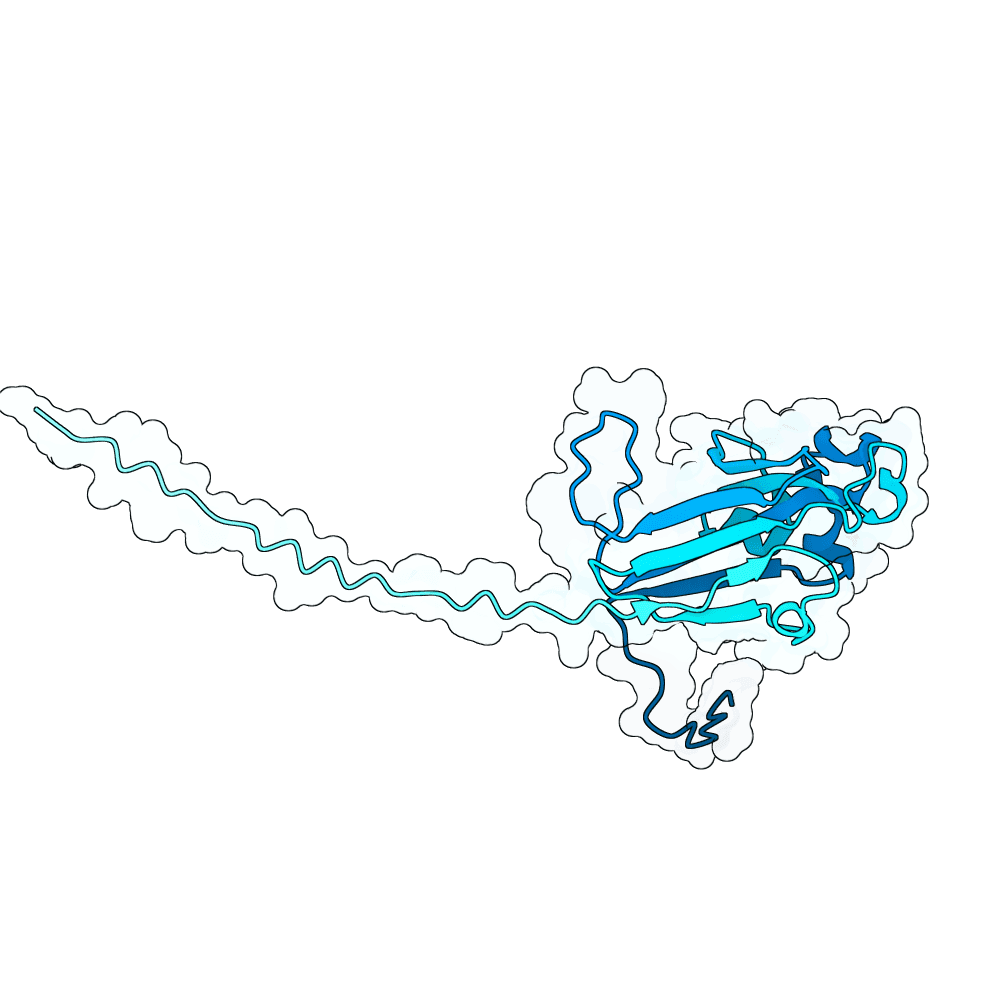
In summary, the PDE generates a very large variant population via diffusion pLMs (Protein Language Models). The population is then scored by several oracles - which are fine-tuned versions of pLMs tailored for specific properties. This scoring process is repeated many times in a genetic algorithm-based optimization process; we typically start with between 100,000 and 1,000,000 variants and iteratively cull them down to 100. For this specific run, we diversified Human Ephrin B2 or B3 into 128,000 protein variants and iteratively culled them to 100 over the course of 100 optimization steps, meaning over 10 million protein variants were generated in total.
Proteins were scored using our oracles that model properties such as PPI (Protein-Protein Interaction) probability (Synteract3), protein-protein binding affinity (Synteract2), foldability, solubility, expressibility, and thermostability, alongside pLM PLL (averaged per-residue log-likelihood) and an internal proxy for "realness." This run also had a heavy emphasis on crossovers in the genetic algorithm, employing a tournament-based approach that chooses parents with a likelihood based on their composite fitness. This helps the population hone in on valuable protein regions. Parents are kept when crossovers are employed, allowing the total variant population at a given step to get quite large.
Finally, we employ AlphaFold3 look-alikes (like Chai-1 or Boltz-2) to calculate interface-based metrics on the last 100 most promising candidates, including iPTM, ipSAE, pdockq, pdockq2, and LIS, which are used to construct our final submission list. We more heavily weighted ipSAE than normal due to it being the competition’s chosen metric. We plan on releasing Synteract3 and the PDE in the first quarter of 2026.