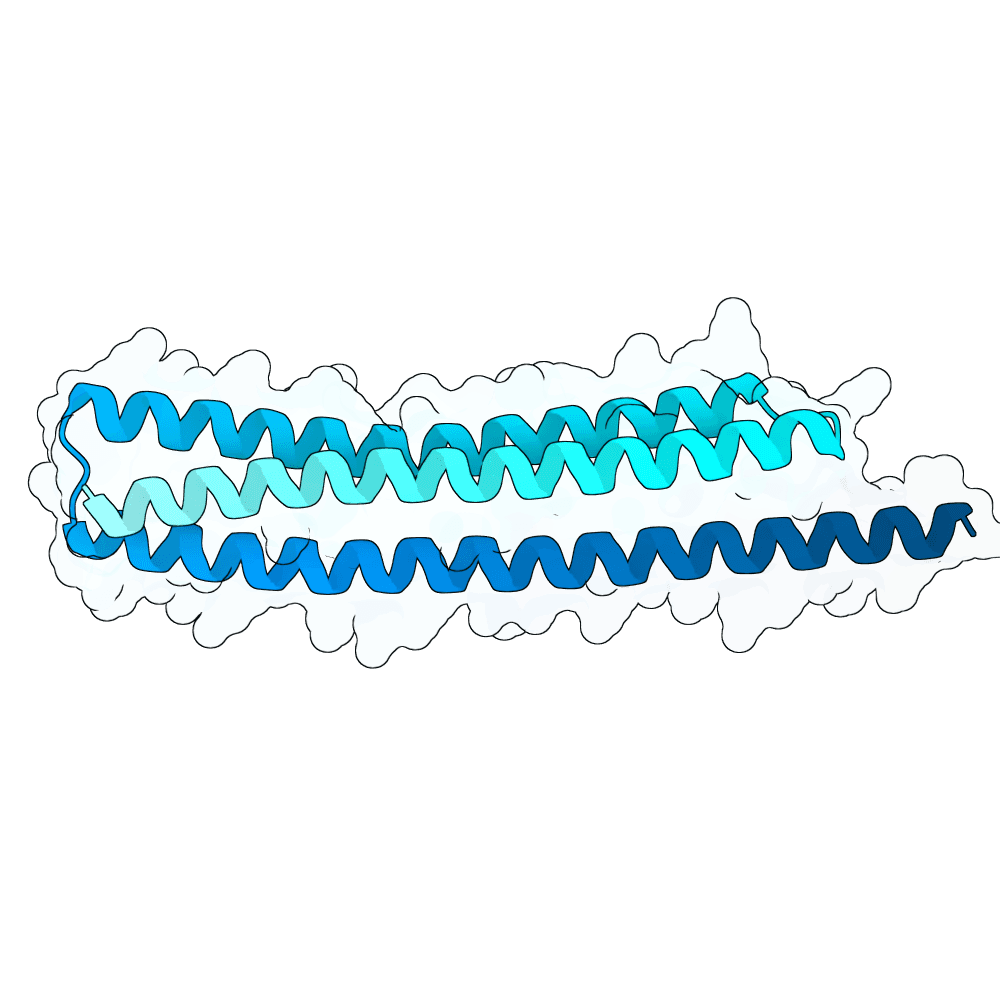
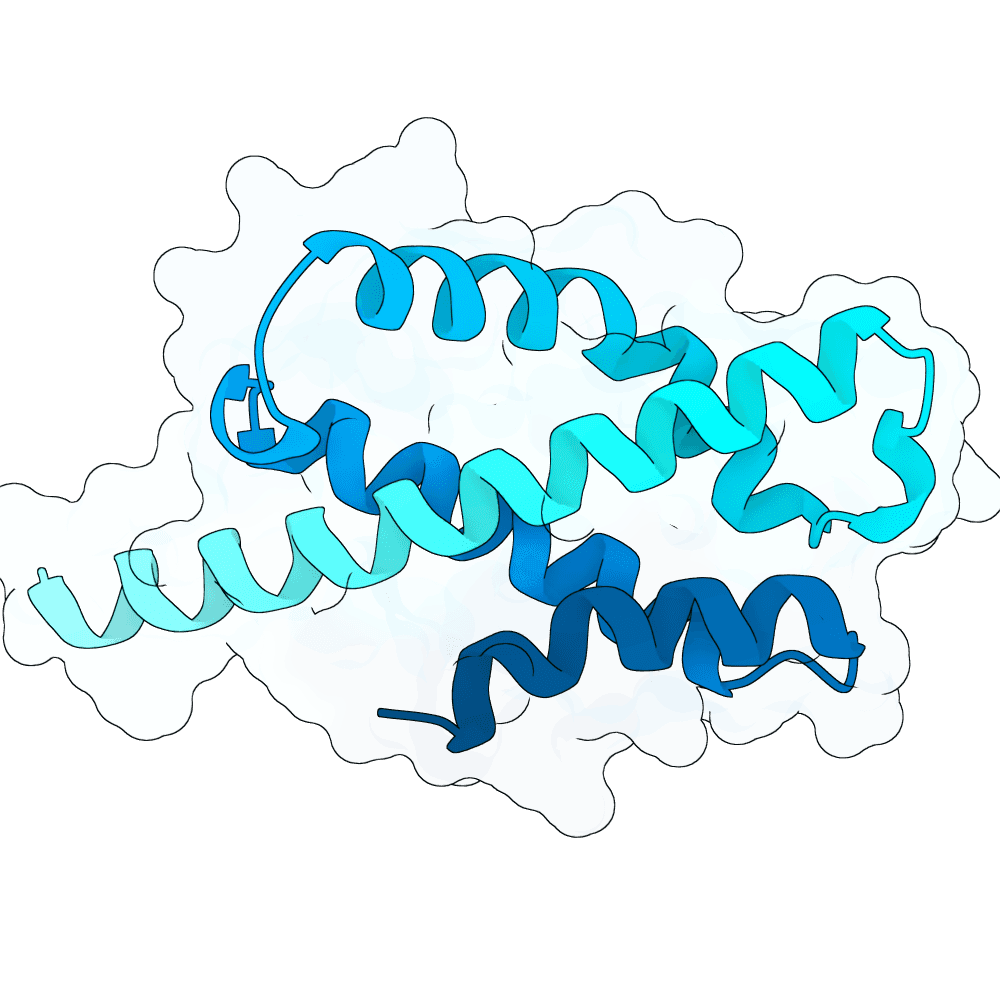
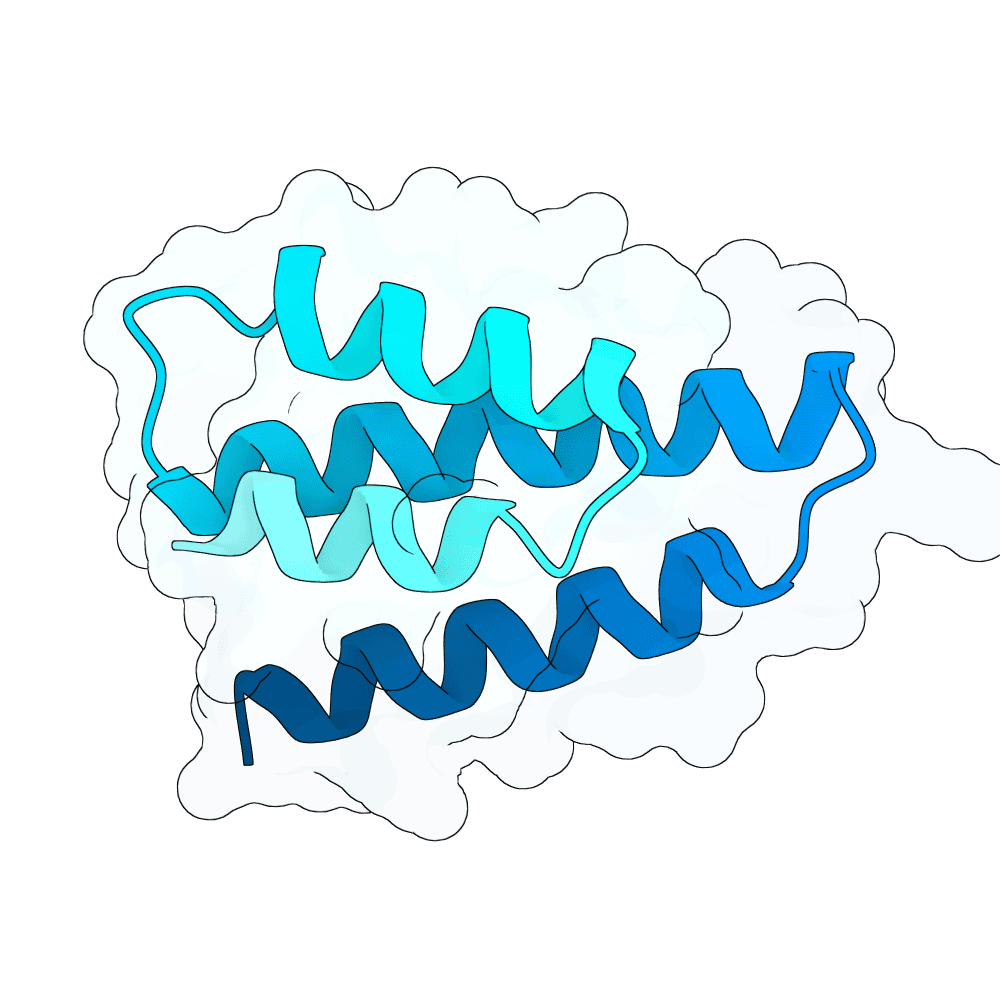
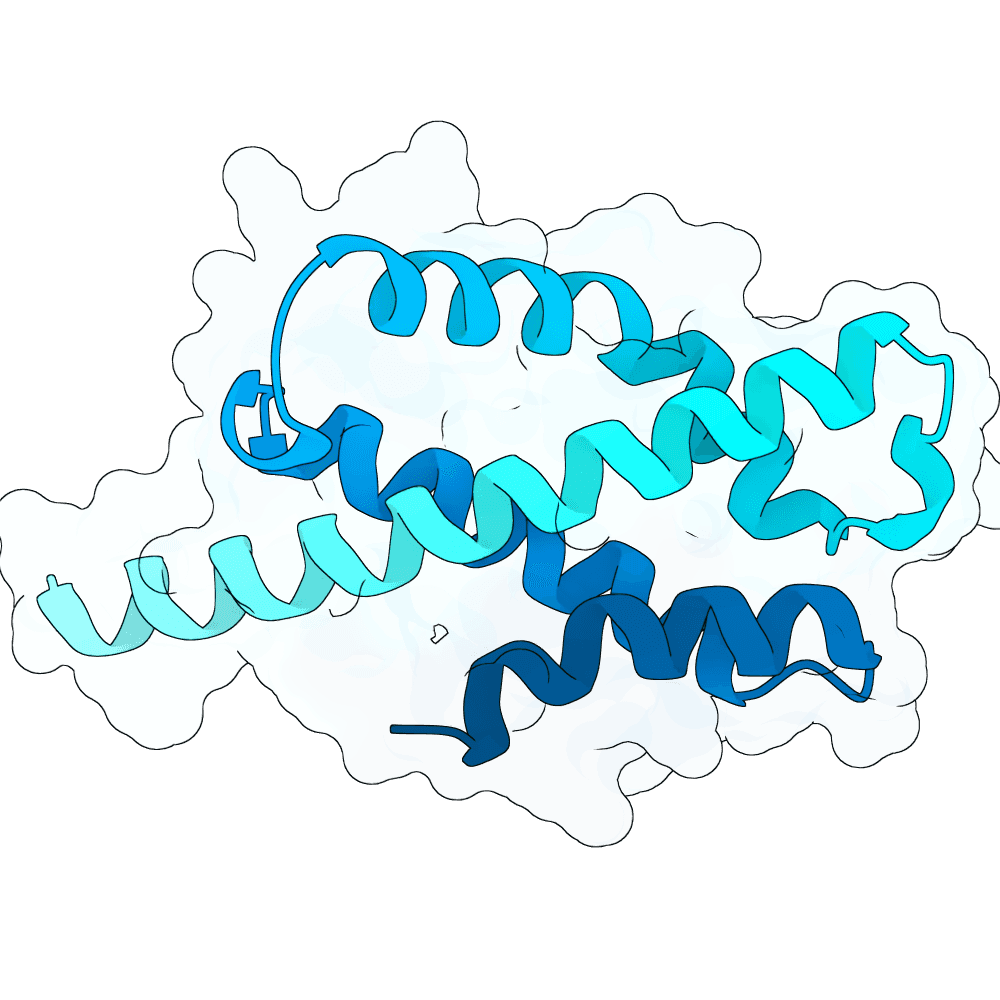
Description
BindCraft is a computationally expensive process. Some trajectories passed initial stage with high iptm scores but were rejected from the filter after MPNN optimisation. We attempted to use Soluble Caliby, a Potts model based sequence design method, (Shuai et al., 2025) to rescue those designs. Interface residues predicted by BindCraft was fixed. Collab effort from Gabriel Ong and Honghao Su.