Description
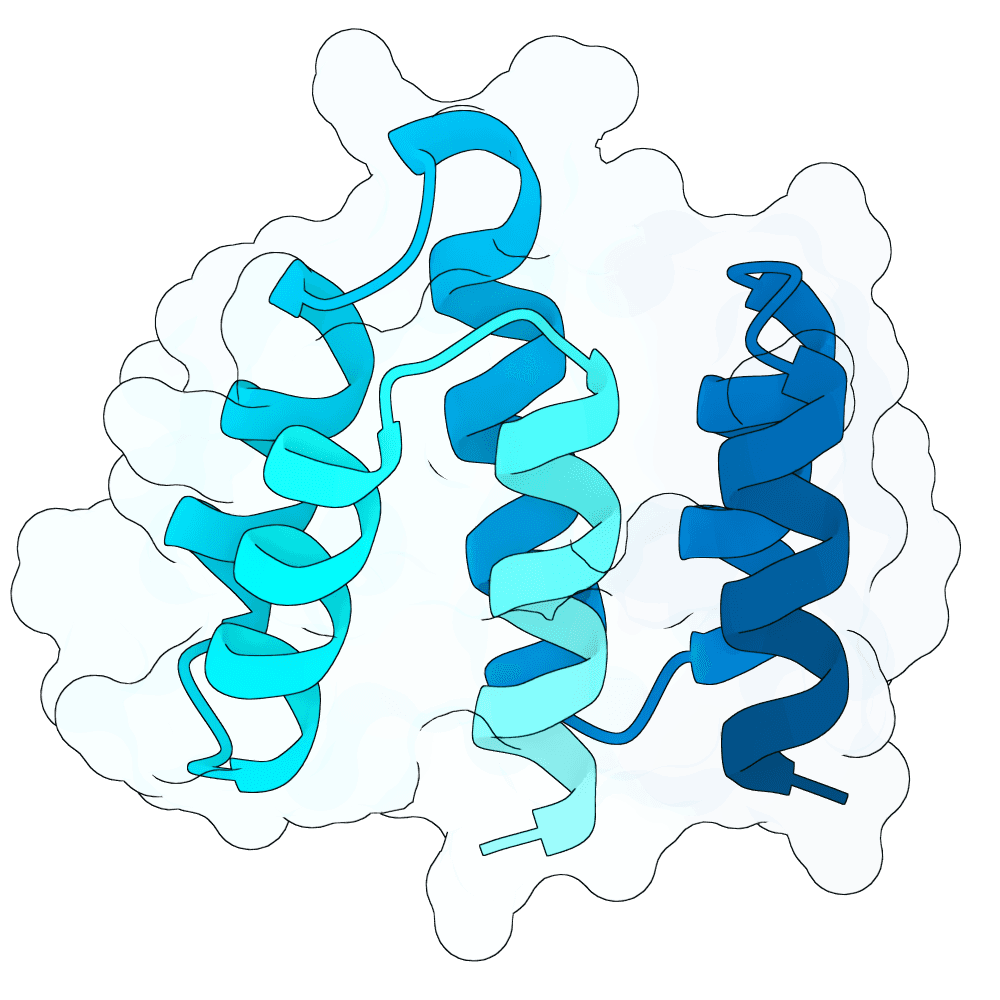
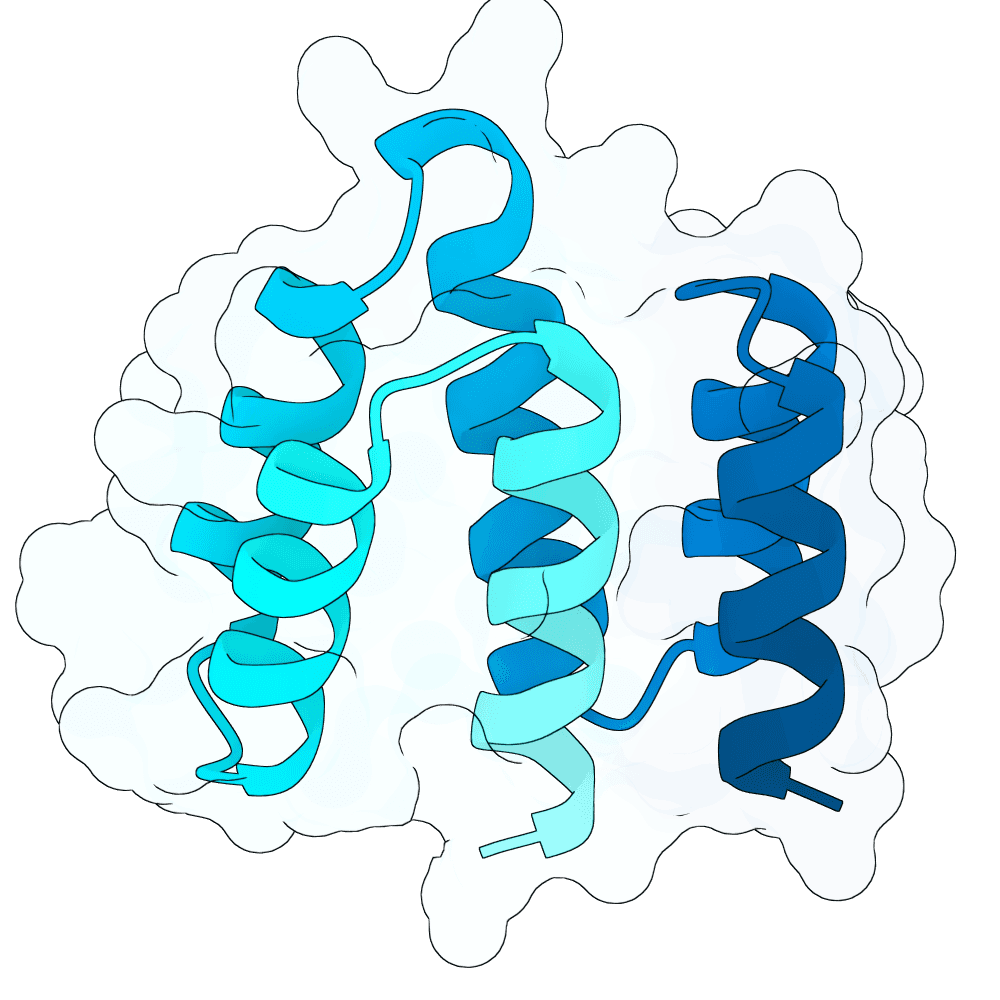
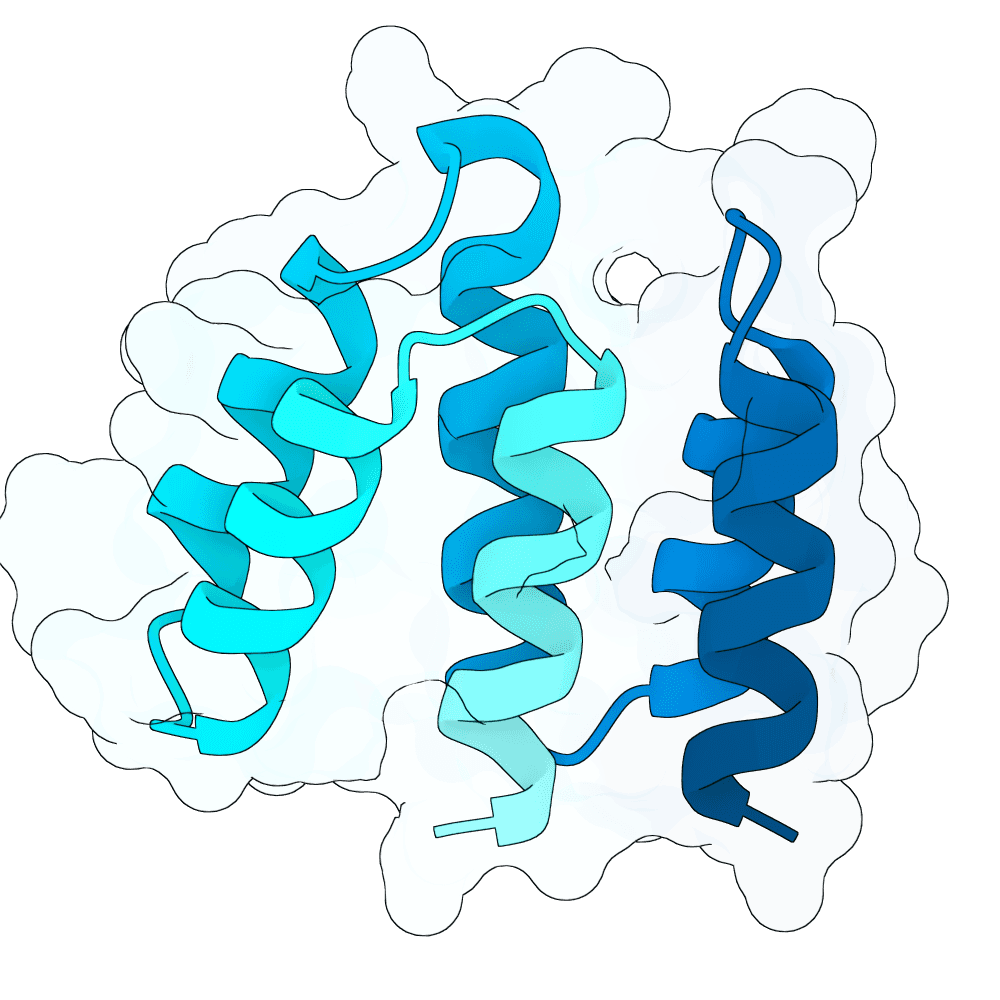
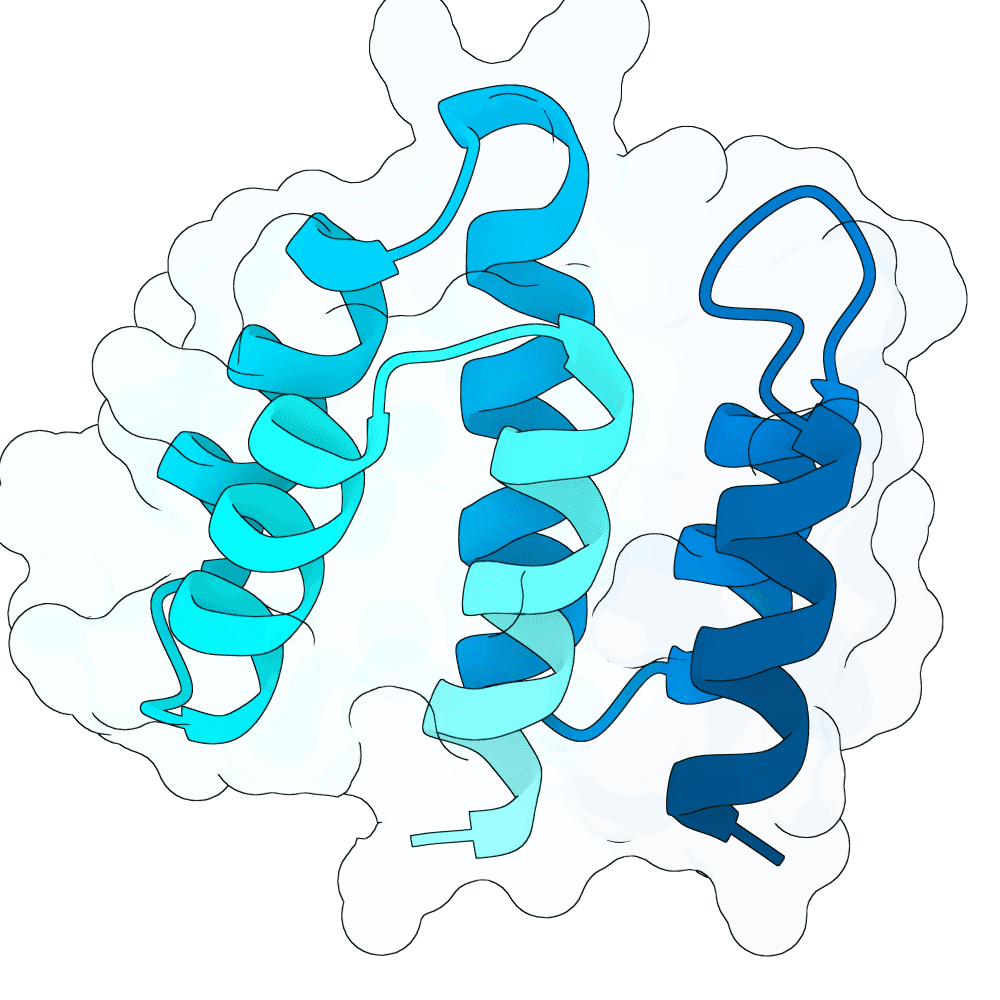
These designs are from the 'vanilla' BoltzGen protein-anything protocol, 65 - 120 aa binders, from 4000 designs with budget of 40, default filters, as implemented in the nf-binder-design pipeline for more efficient parallelism on HPC. I used two hotspots W504 and Y581 near the Ephrin binding site, set the high B-factor loops to 'visibility: 0' to encourage flexibility and added not_binding 'anti-hotspots' in a band around the middle of the protein to reduce off-target designs.
I refolded the 40 final designs with Boltz-2 to calculate ipSAE_min for each and took the best scoring representative (the main branch of BoltzGen doesn't output a PAE.npz for ipSAE yet, hence the need to re-fold this way - probably in a few weeks BoltzGen will output ipSAE directly).
Additionally with that best representative I elaborated a small loop between two helices with 1..4 additional residues that looked like it could be extended to make better contacts (using BoltzGen design_insertions, 200 designs x3 inverse folding sequences). I then selected the top three loop-elaborated sequences by ipSAE_min. These have all very similar ipSEA_min - will be interesting to see the impact on affinity.