Proteins (8)
id: quiet-orca-jade
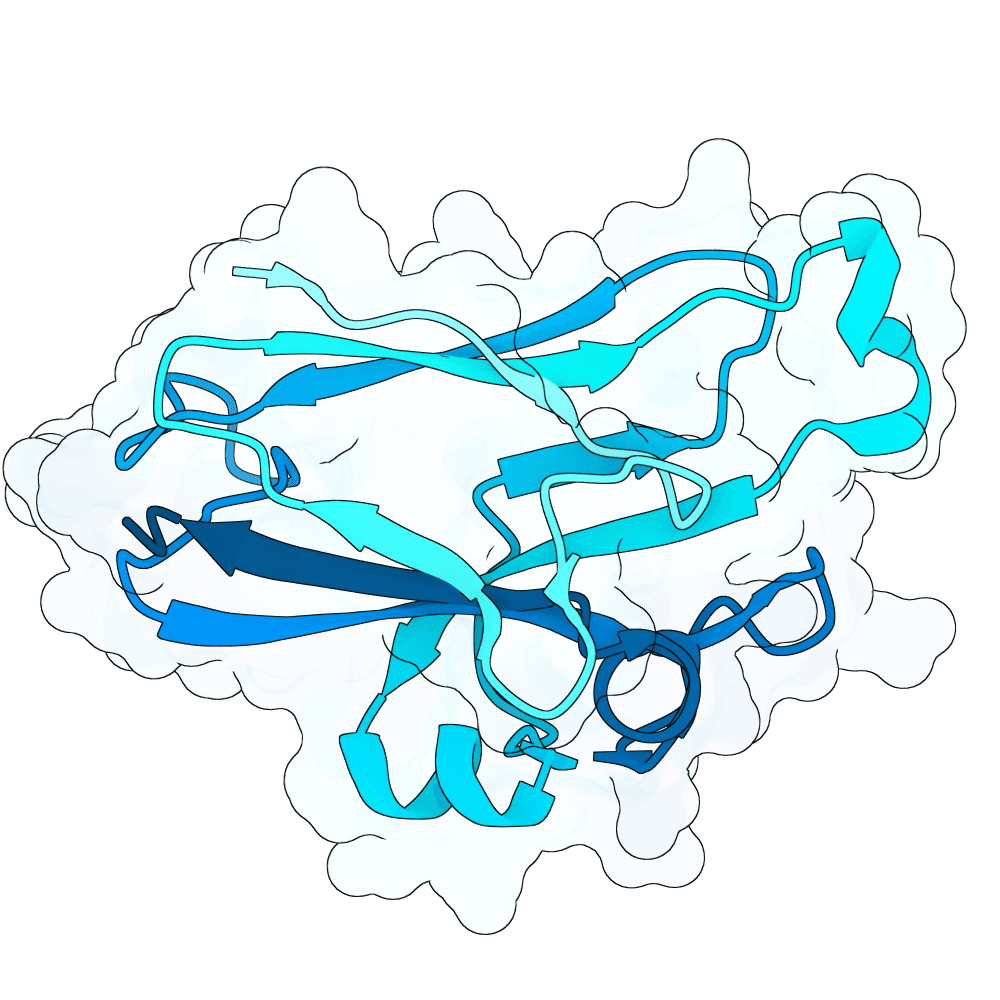
Nipah Virus Glycoprotein G
None
81.93
True
16.0 kDa
140
id: quiet-heron-rose
Nipah Virus Glycoprotein G
None
81.38
True
15.9 kDa
140
id: silent-lynx-lotus
Nipah Virus Glycoprotein G
None
80.95
True
16.0 kDa
140
id: strong-mole-cloud
Nipah Virus Glycoprotein G
None
81.42
True
15.9 kDa
140
id: brisk-kiwi-snow
Nipah Virus Glycoprotein G
None
81.68
True
15.9 kDa
140
id: radiant-seal-opal
Nipah Virus Glycoprotein G
None
81.96
False
15.9 kDa
140
id: wild-mole-iron
Nipah Virus Glycoprotein G
None
81.12
False
15.9 kDa
140
id: green-eagle-maple
Nipah Virus Glycoprotein G
0.77
81.80
--
16.0 kDa
140