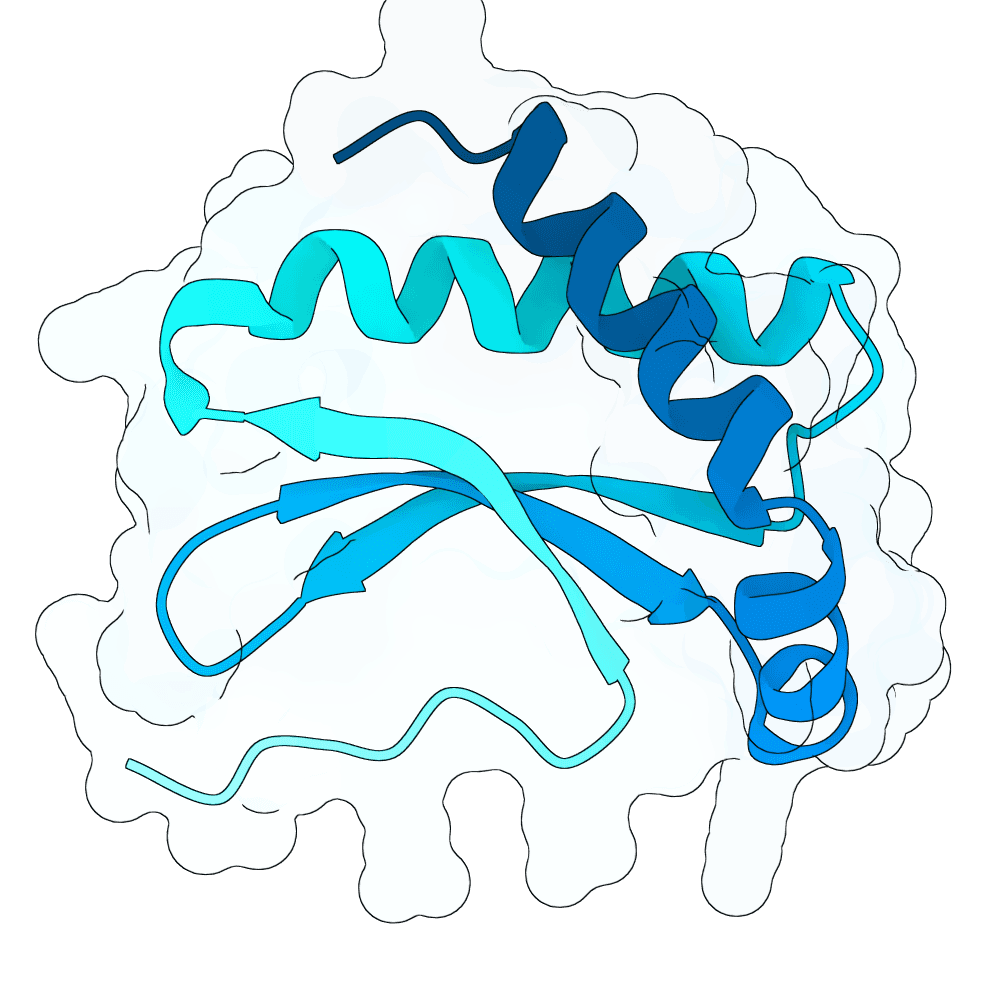
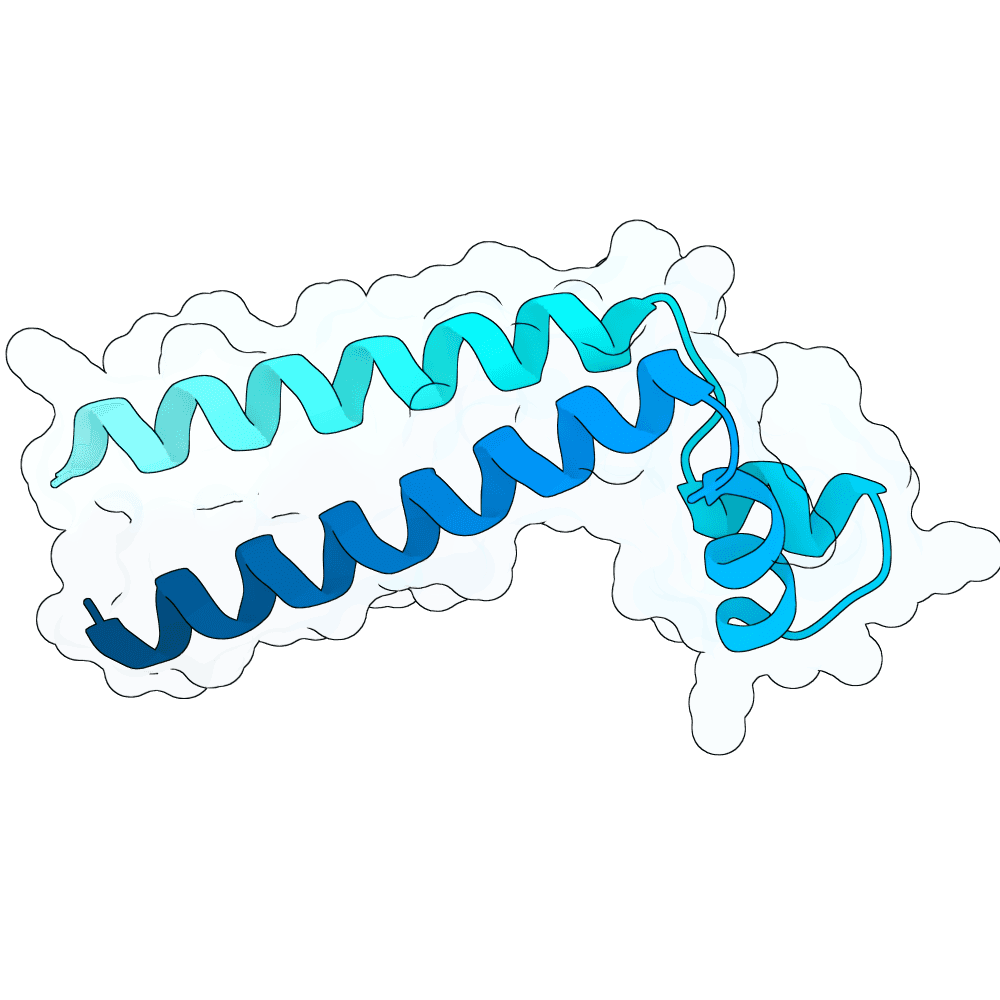
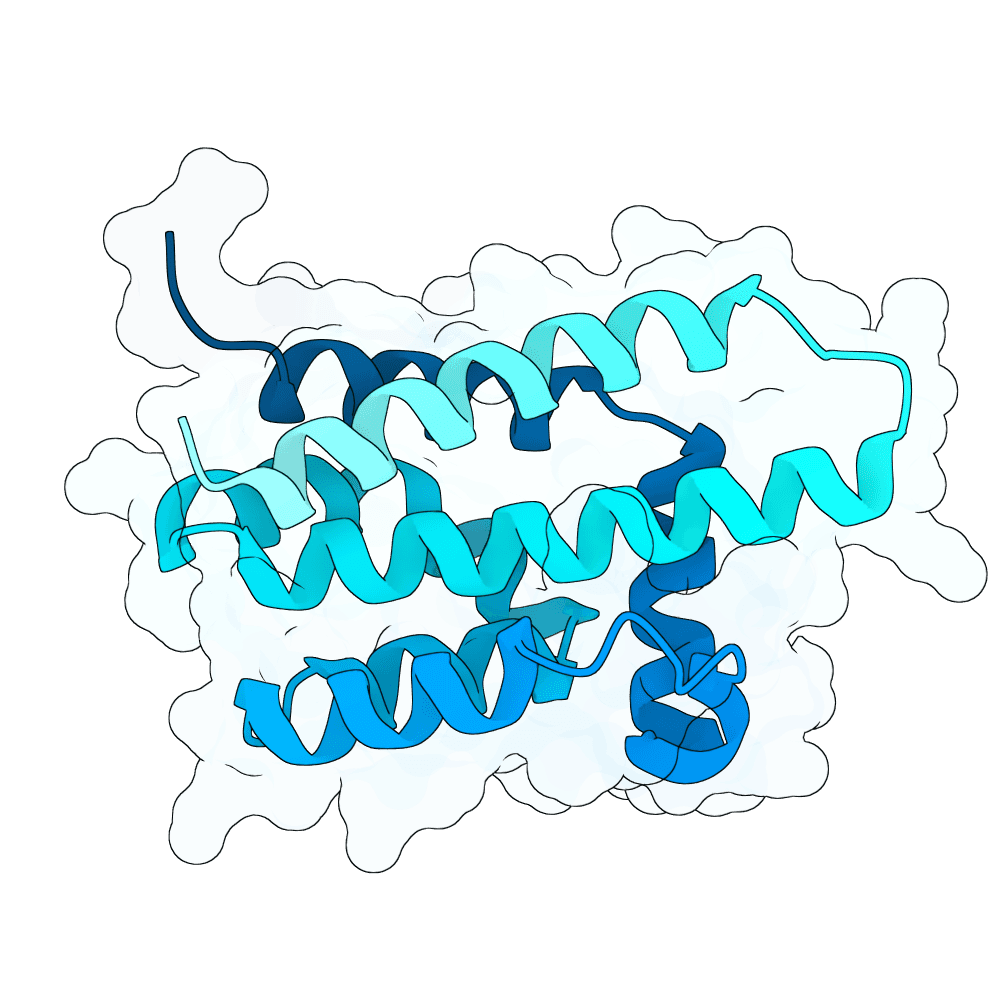Description
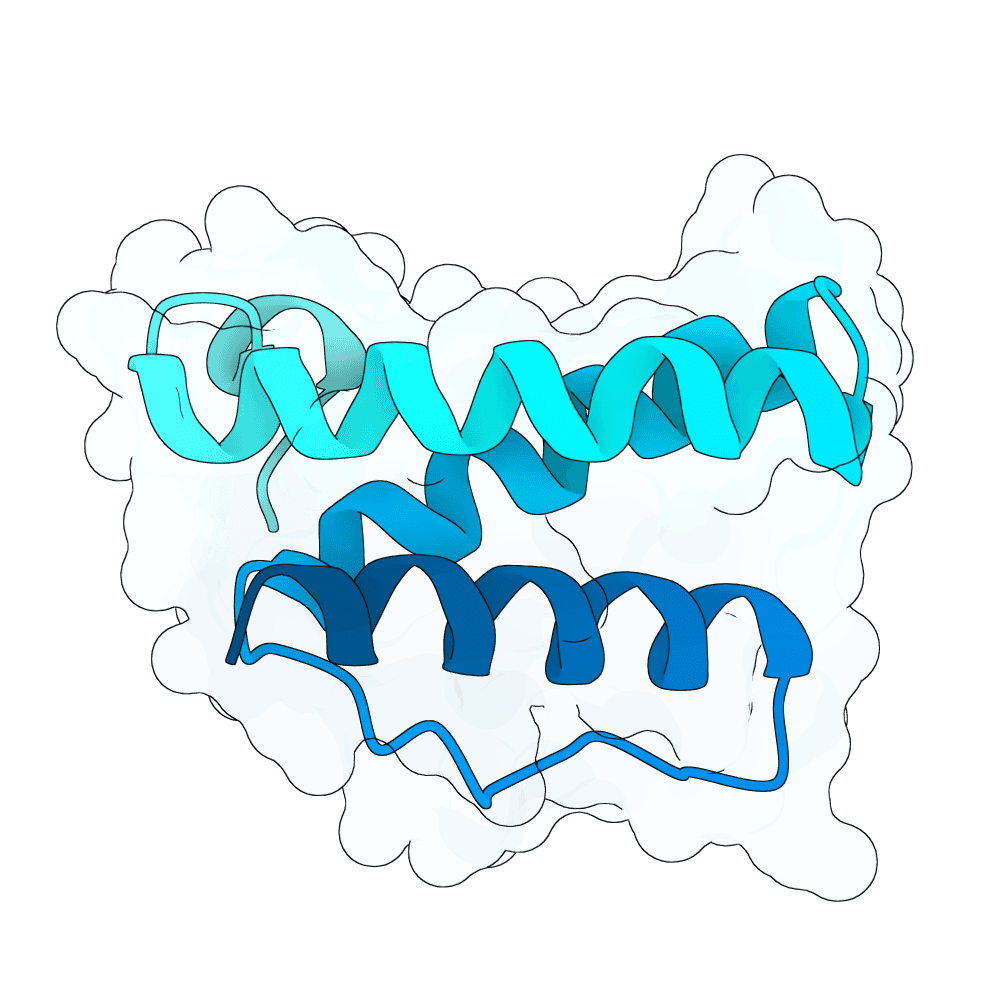
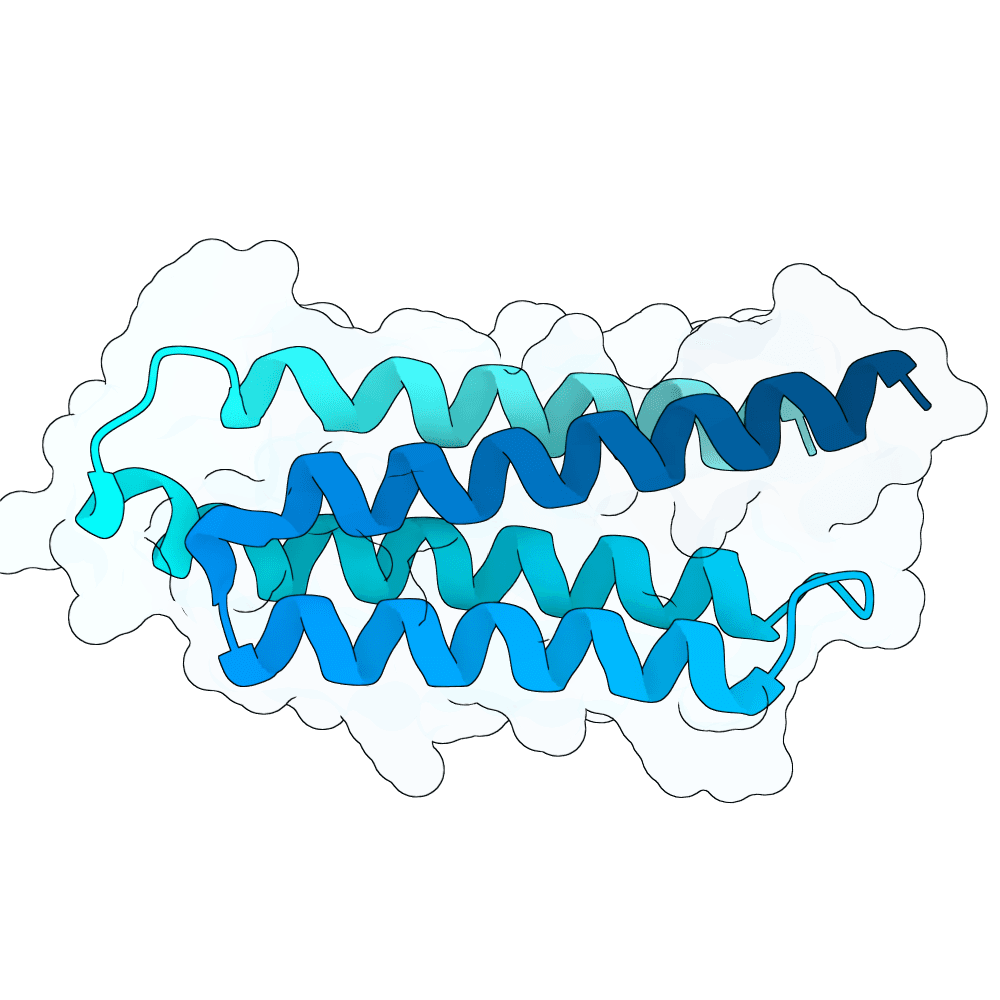
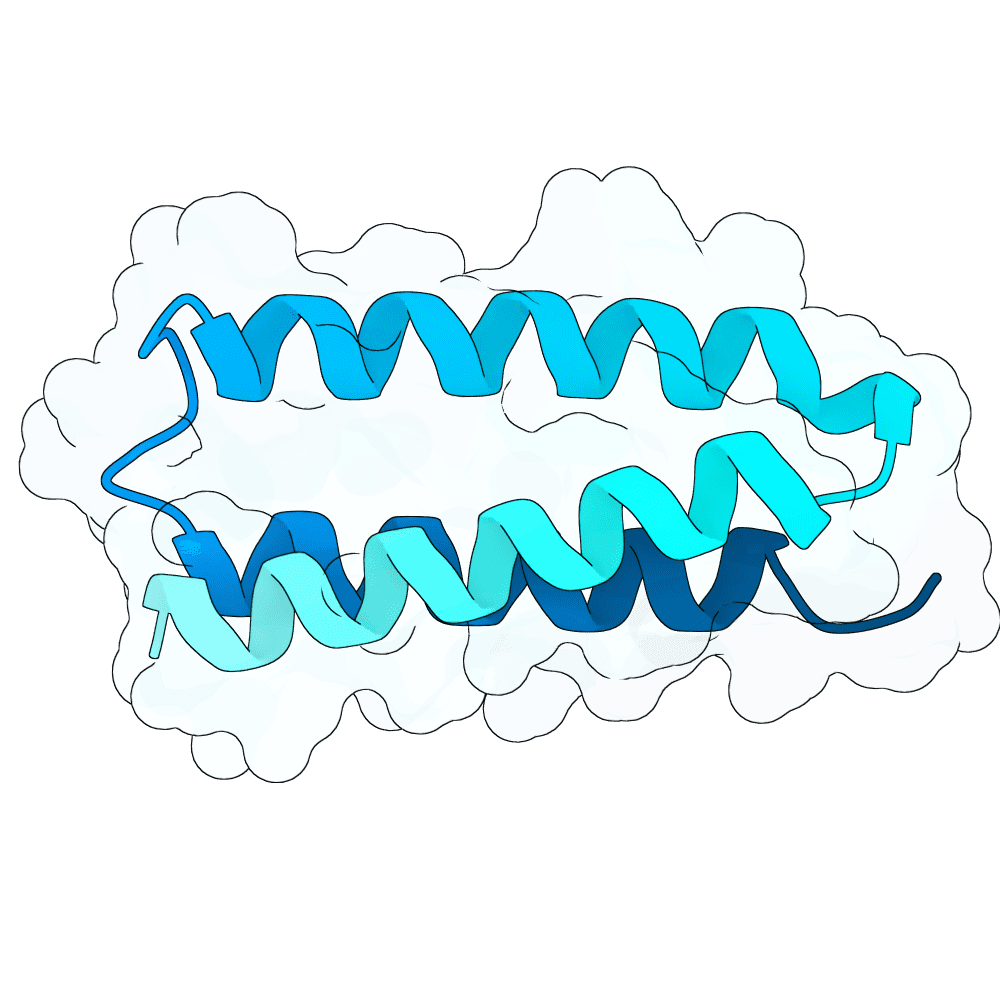
All submitted protein sequences are de novo designs generated with the BindCraft pipeline using the default_4stage_multimer_hardtarget protocol. Hotspots were chosen based on the binding surface observed in the reference PDB structure, which was trimmed to the minimal binding interface region to reduce memory requirements during design.