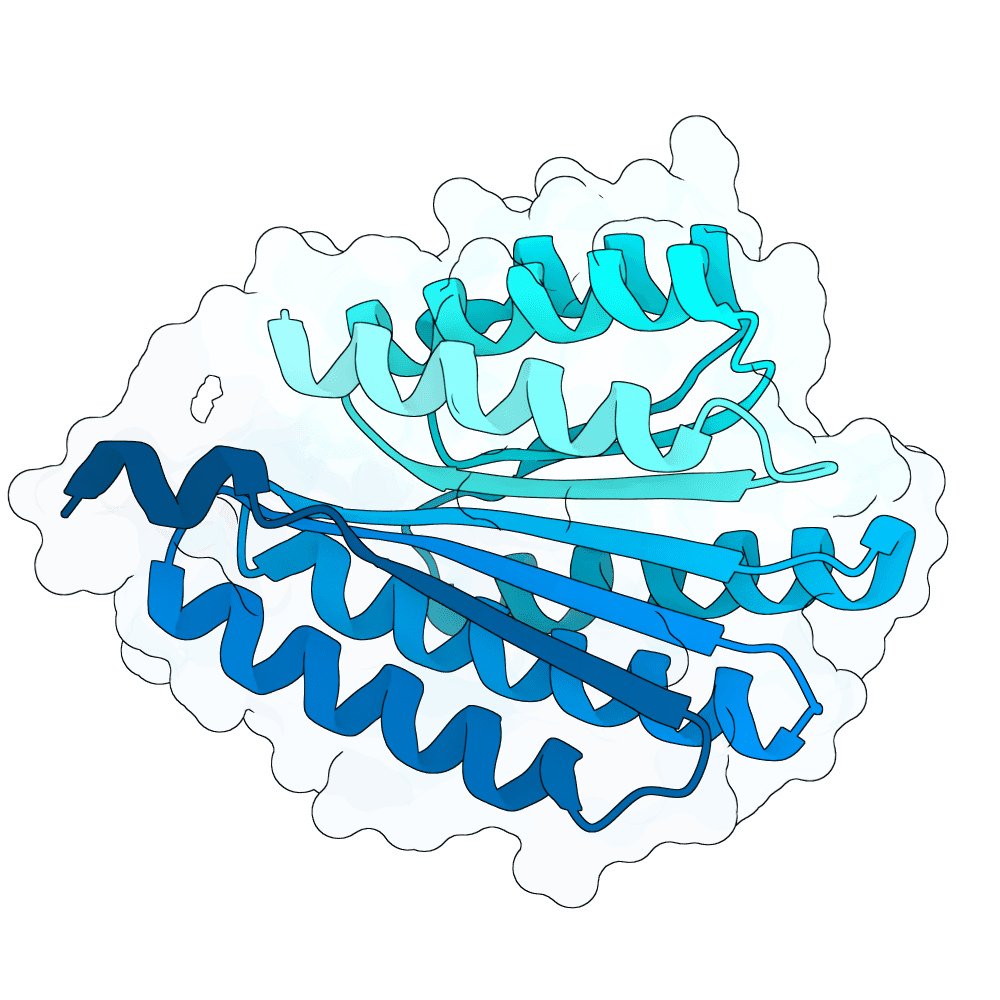
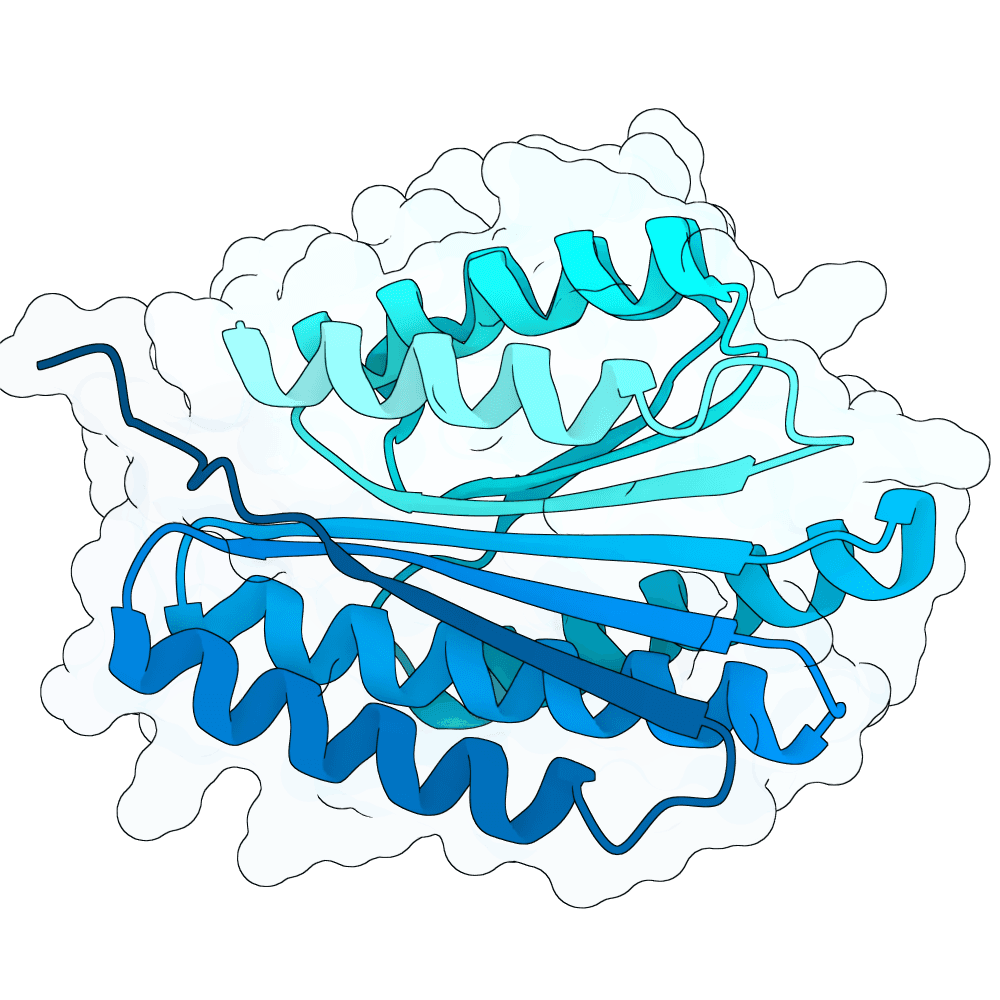
Submitted toNipah Binder Competition
Description
Began with Rf diffusion to generate backbones and chose a promising helical binder structure. Used this structure to run protein MPNN and generate sequences. Refolded using AlphaFold3 first, followed by Boltz2. Ultimately used Boltz2 to choose best sequences.