Description
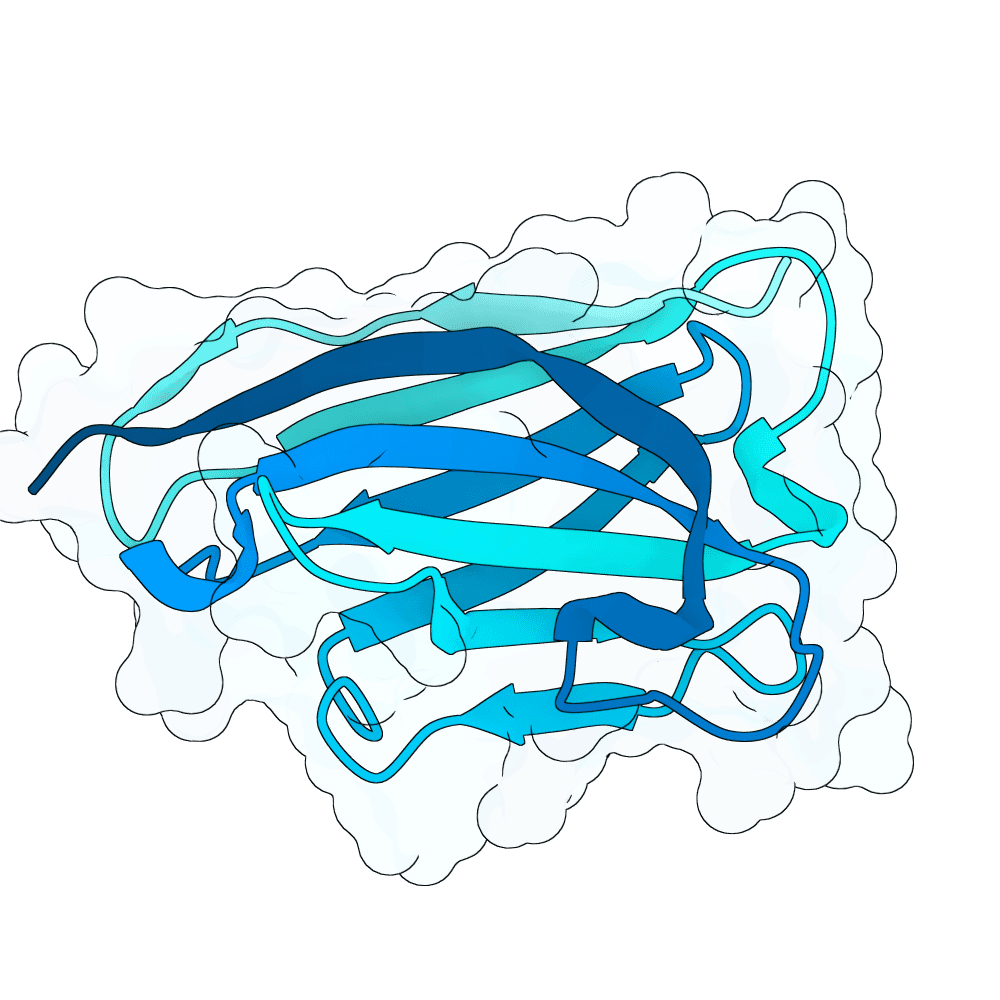
Binder Design Design aim: We aimed to design nanobody-based binders with neutralising activity against the Nipah Virus, focusing on disrupting the NiV-G / EFNB2 interaction.
Hotspot ID:
- Using the 2vsm structure from Adaptyv, BioPython was used to identify EFNB2-contacting residues (defined as carbon atoms within 10 Å of the opposing chain).
- Additional hotspot positions were selected based on DMS data from Larson et al. (Cell, 2025).
- A PyMol analysis was performed to identify a small hydrophobic patch suitable for targeting.
- Final hotspot residues used for design: I401, R402, W504.
Sequence Similarity Network Analysis:
- A Sequence Similarity Network (SSN) was built to evaluate diversity across CDR1, CDR2, and CDR3.
- Levenshtein distance matrices were computed using
stringdist(method = "lv"), and distance thresholds were applied to define network edges. - Networks were constructed with igraph, allowing identification of clusters and highly similar sequence families.
- iPTM-based metrics were mapped onto nodes to highlight high-scoring design clusters for both iptm and ipSAE.
- This SSN framework guided the selection of representative nanobody variants for downstream scoring.
Binder Score Calculations
Binder Re-folding:
- All selected binders were re-folded using Boltz2, and the resulting structures and confidence matrices were used for downstream scoring.
ipSAE calculation:
- ipSAE was computed from Boltz2 outputs using the ipSAE package with default settings.