Description
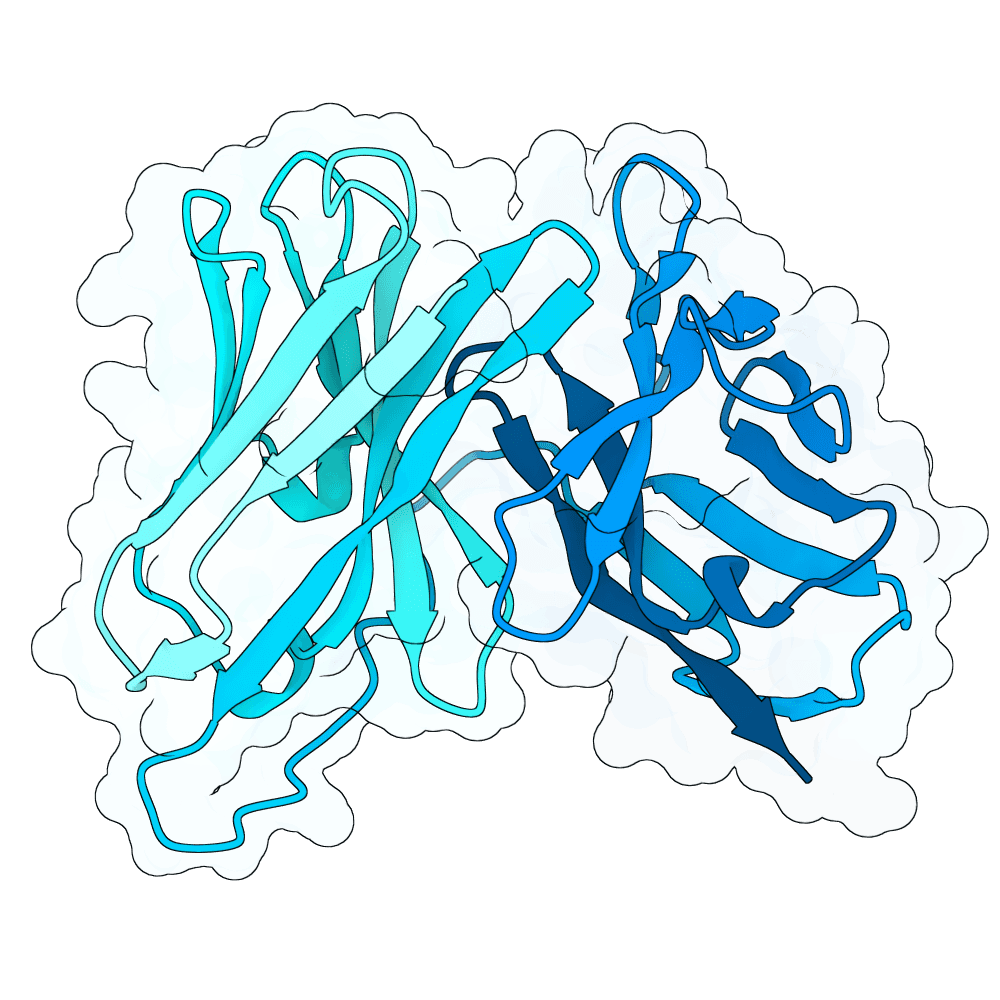
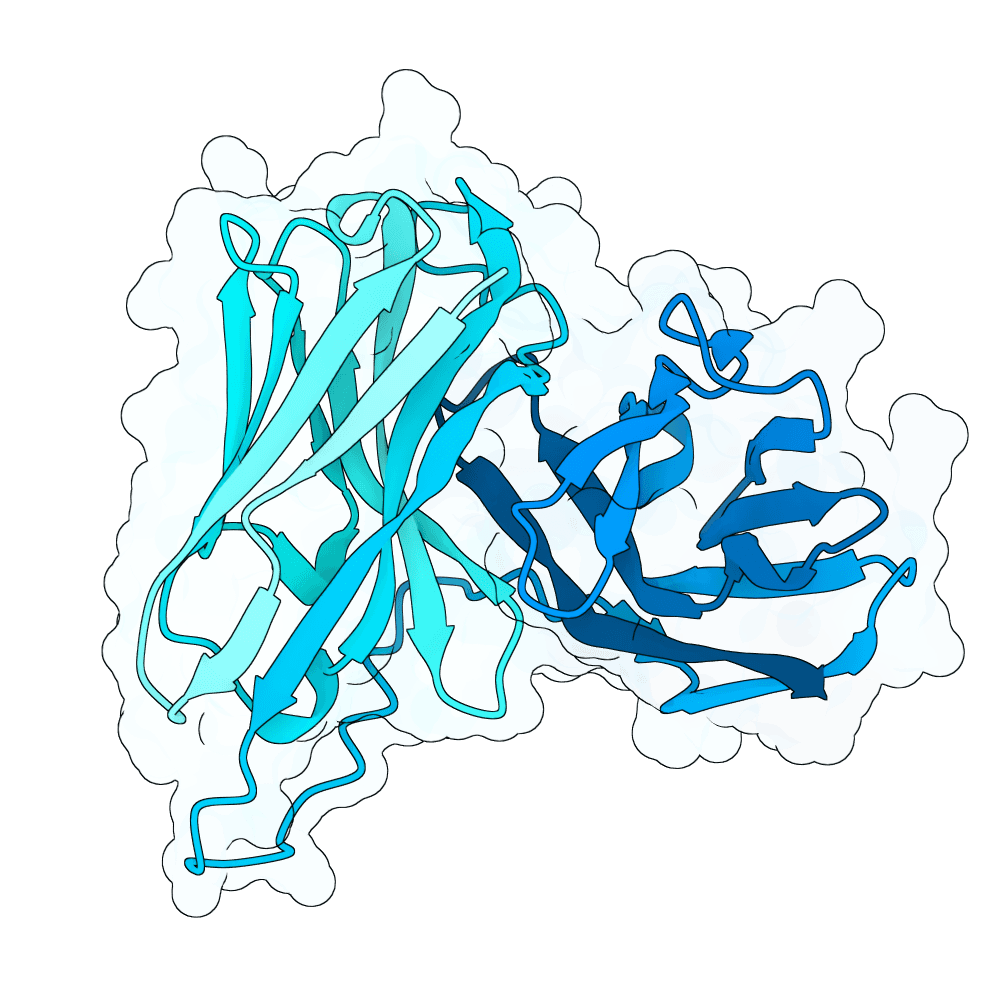
For this competition, I decided to focus on one of the more difficult binder design challenges: de novo design of the antibody CDR-H3 loop. I started with the PDB structures of several mABs that are known binders of NIV and/or HENV, all of which target the same epitope, but in different orientations of each mAB. I converted each mABs to scFV, and used RFDiffusion to apply low noise diffusion to the interface CDR regions, generating 5000 designs.
From there, I followed an iterative protocol that selects one amino acid candidate at a time by maximizing the likelihood between ProteinMPNN and the AntiBERTy language model's output logits. This iterative aa selection process is similar to the protocol proposed by Del Alamo et al at GSK earlier this year.
I used AntiFold to rank the sequences by likelihood given their RFDiffusion-designed backbones. I selected 10 of the top ranked results, with an emphasis on diversity in sequence and predicted scFV orientation w/r/t the target epitope. The selected designs were confidence-checked for feasibility using AF3.