Description
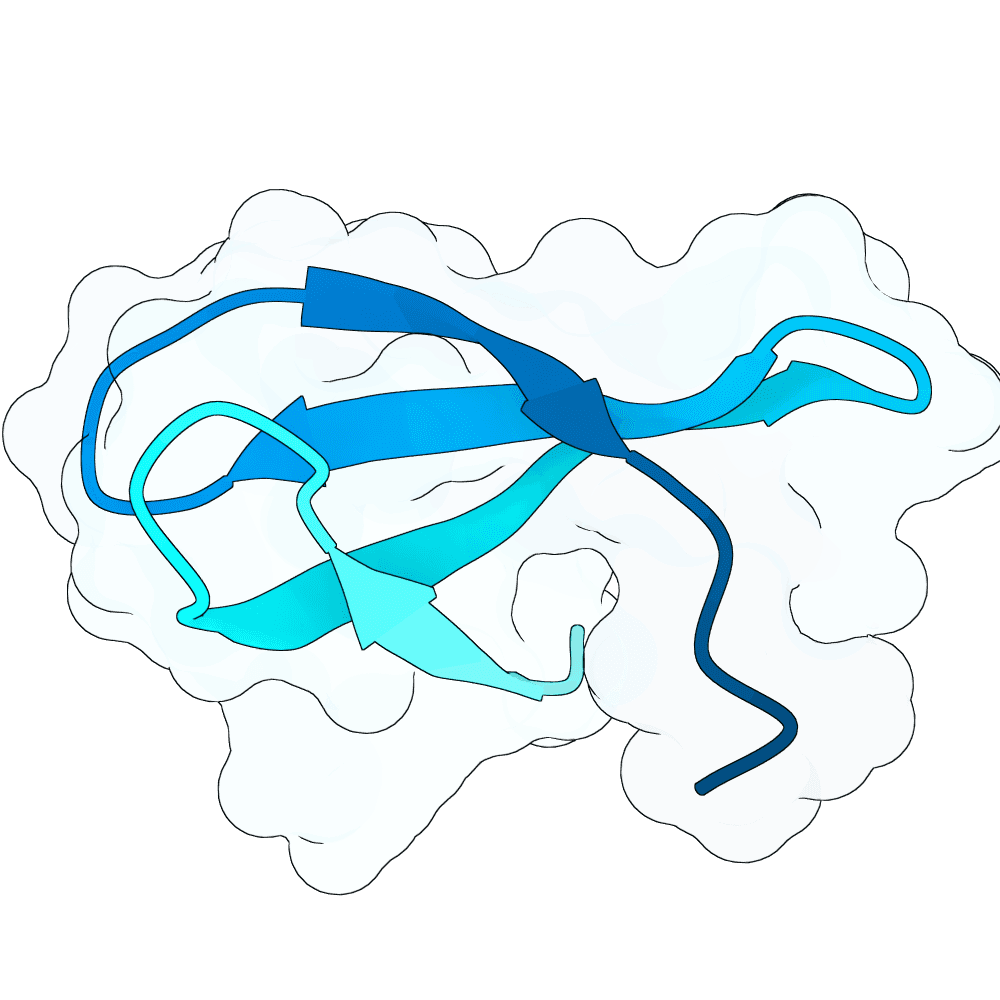
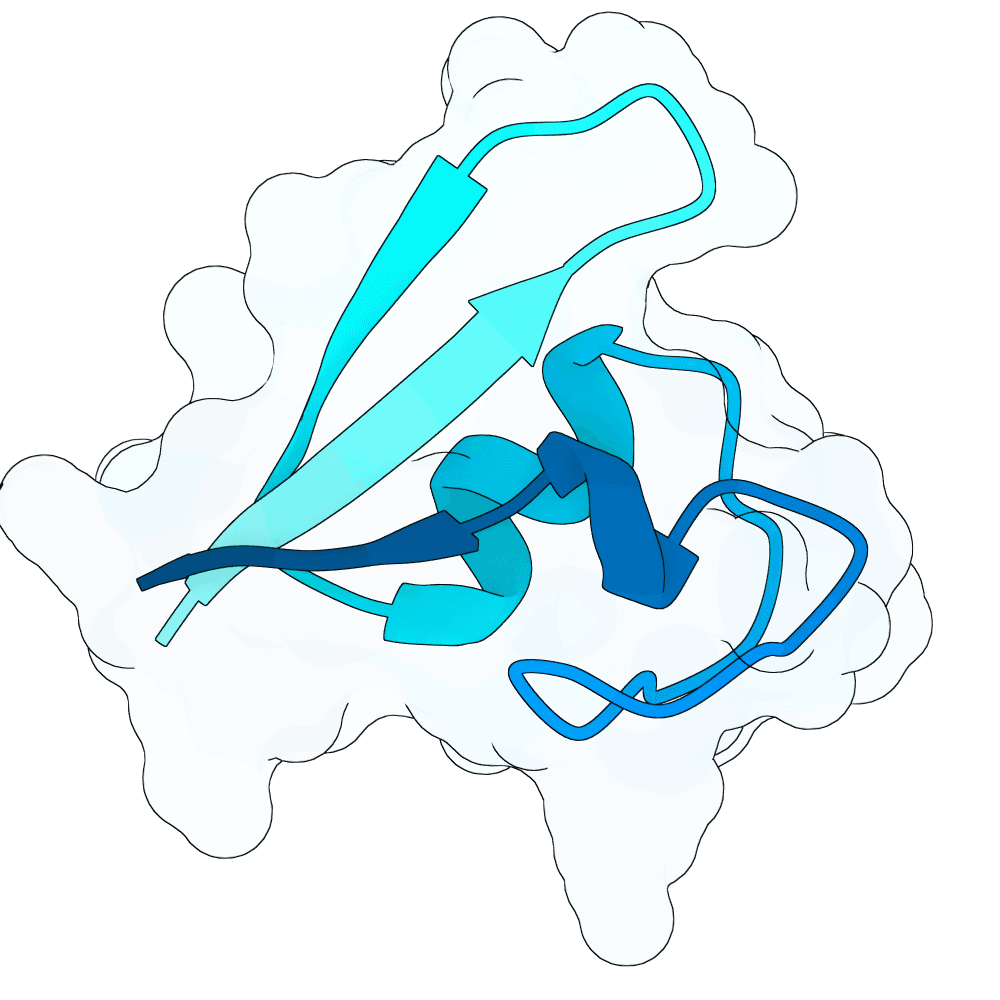
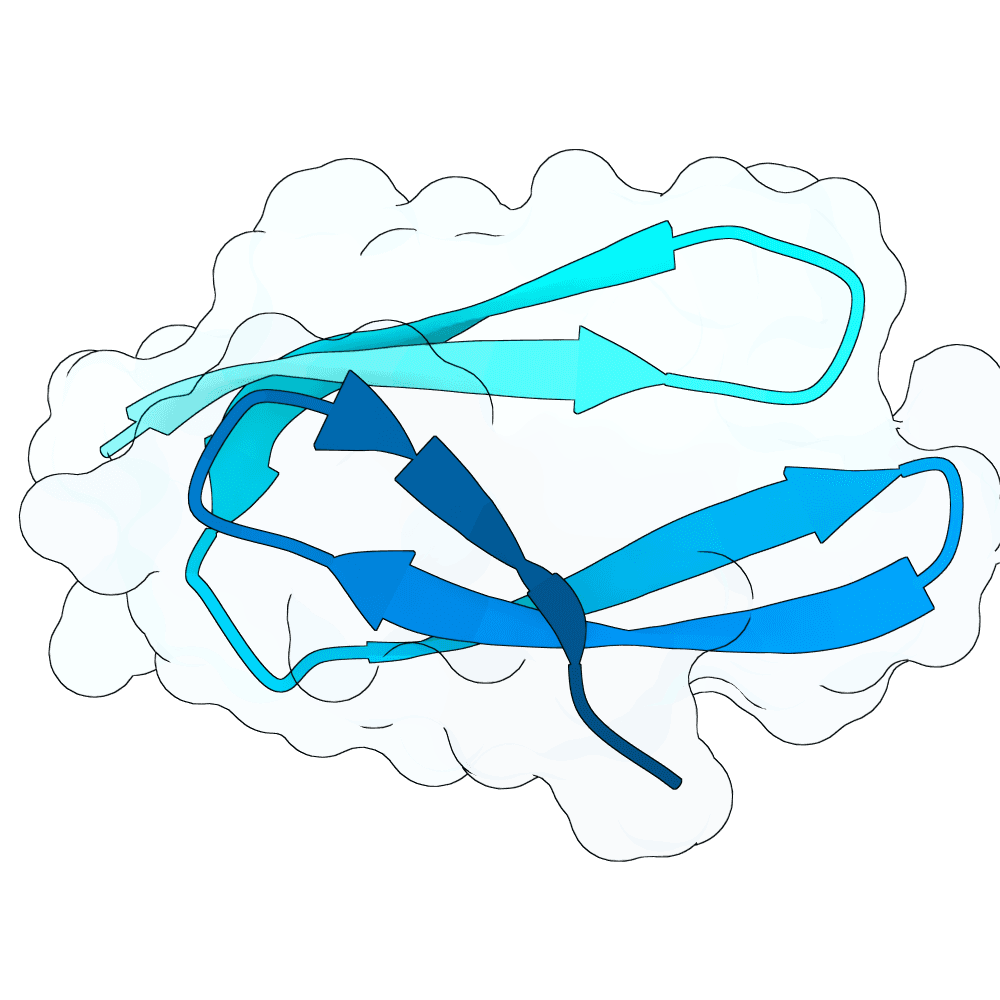
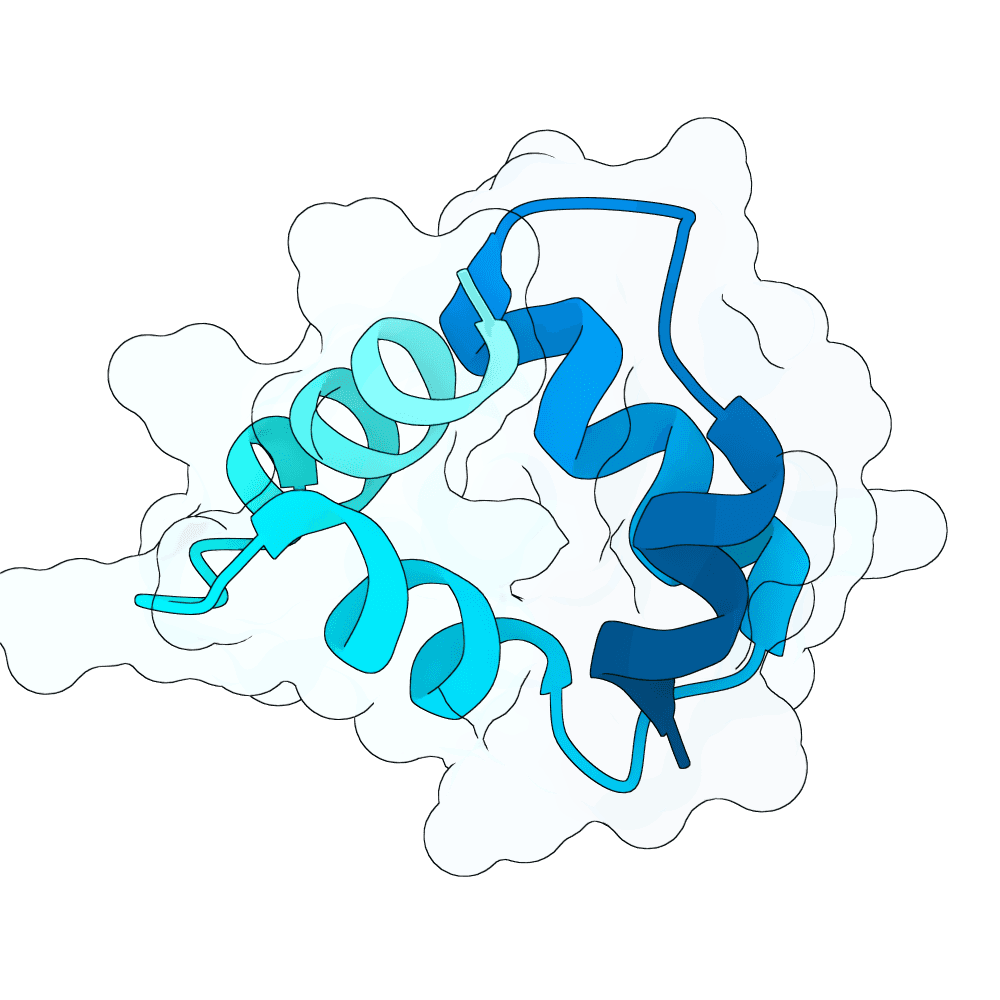
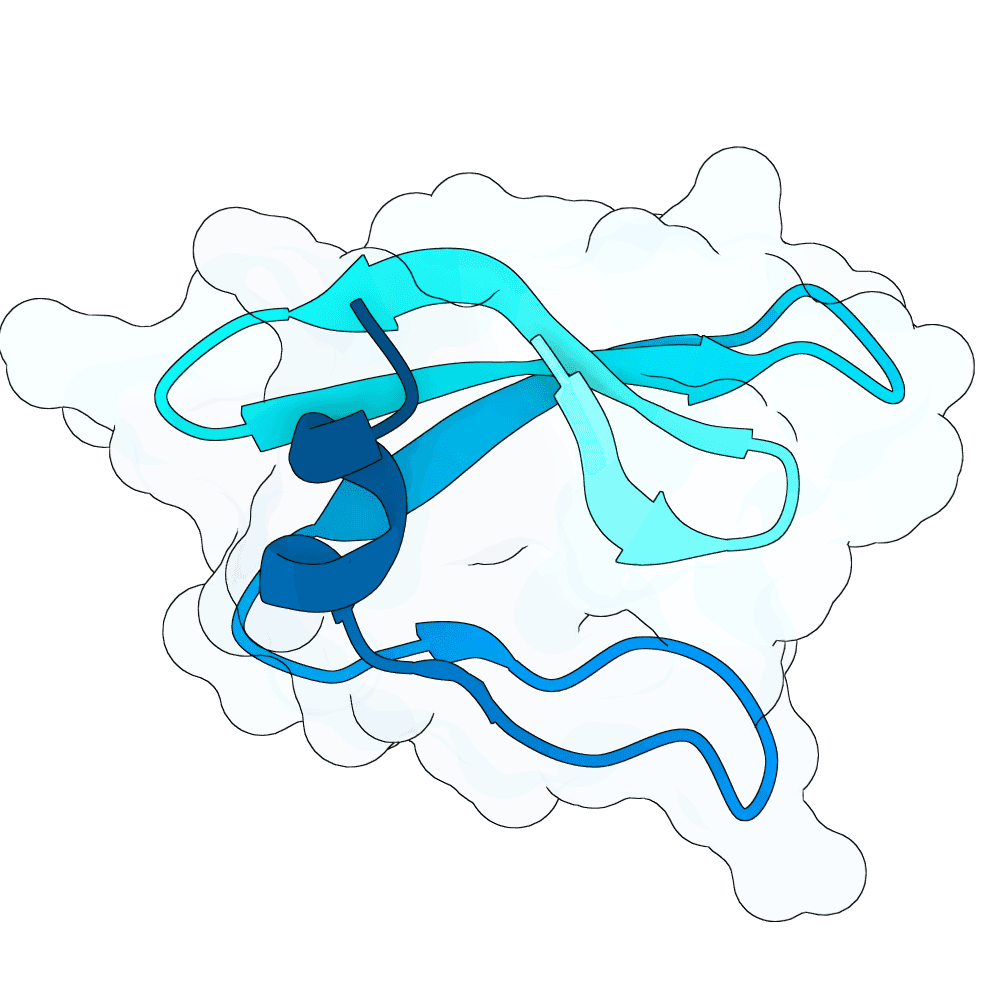
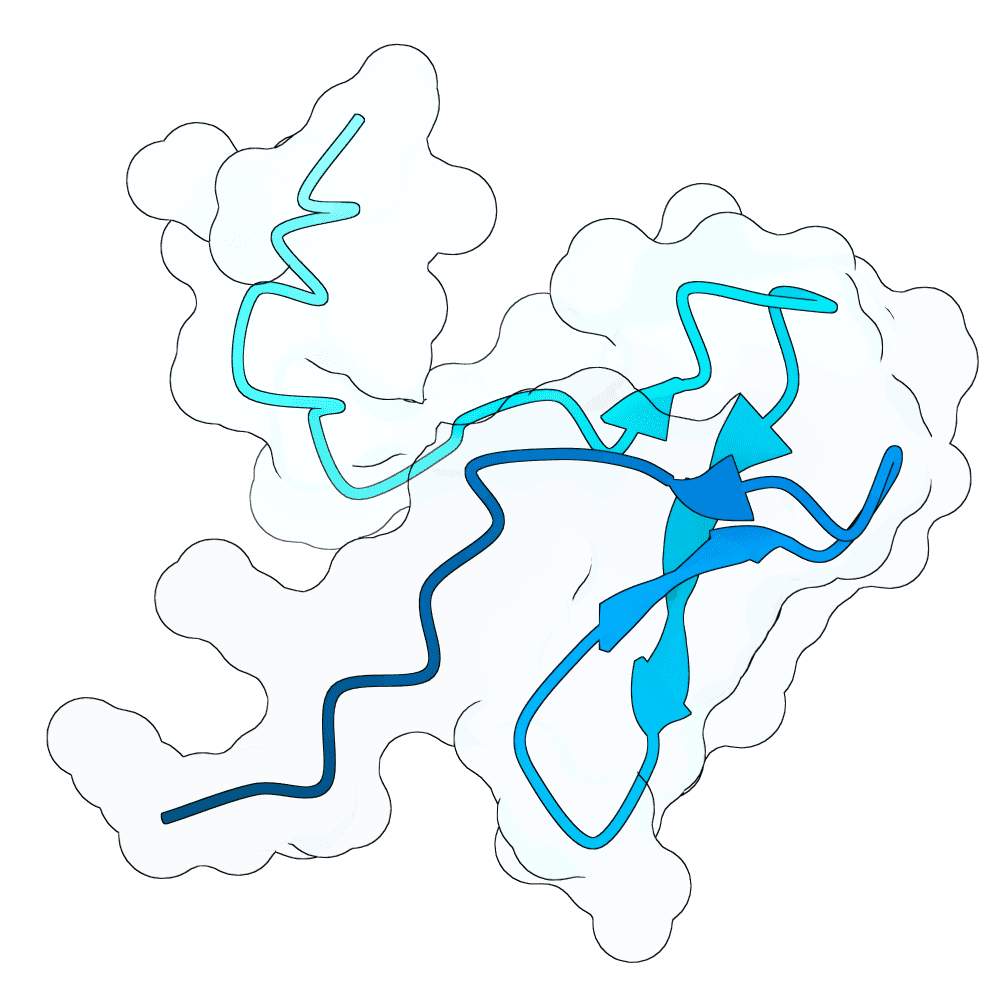
To propose new binders, I used the newly released BoltzGen as a generative prior to sample de-novo short protein sequences (target length: 40–60 AA). The point was not sequence editing of known binders, but generative sampling: BoltzGen was used to draw candidate sequences that the model judges to lie in foldable / protein-like manifold regions of sequence space.
For the top scoring sequences, they were then passed through AlphaFold structure prediction to quickly sanity-check whether the candidate sequences appear to form a compact, stable single-chain topology. Predicted models that were fully or partly disordered were filtered out. Also a BSA analysis in pymol was done.
For the subset of sequences with coherent predicted folds, I then computationally estimated interface scores against the G protein surface using PRODIGY. These interface scores were only used comparatively (to triage candidates), not as absolute physical numbers.
Two of the top-scoring candidates (sequence 9 and 10 in our set) were intentionally generated as cyclic sequences.