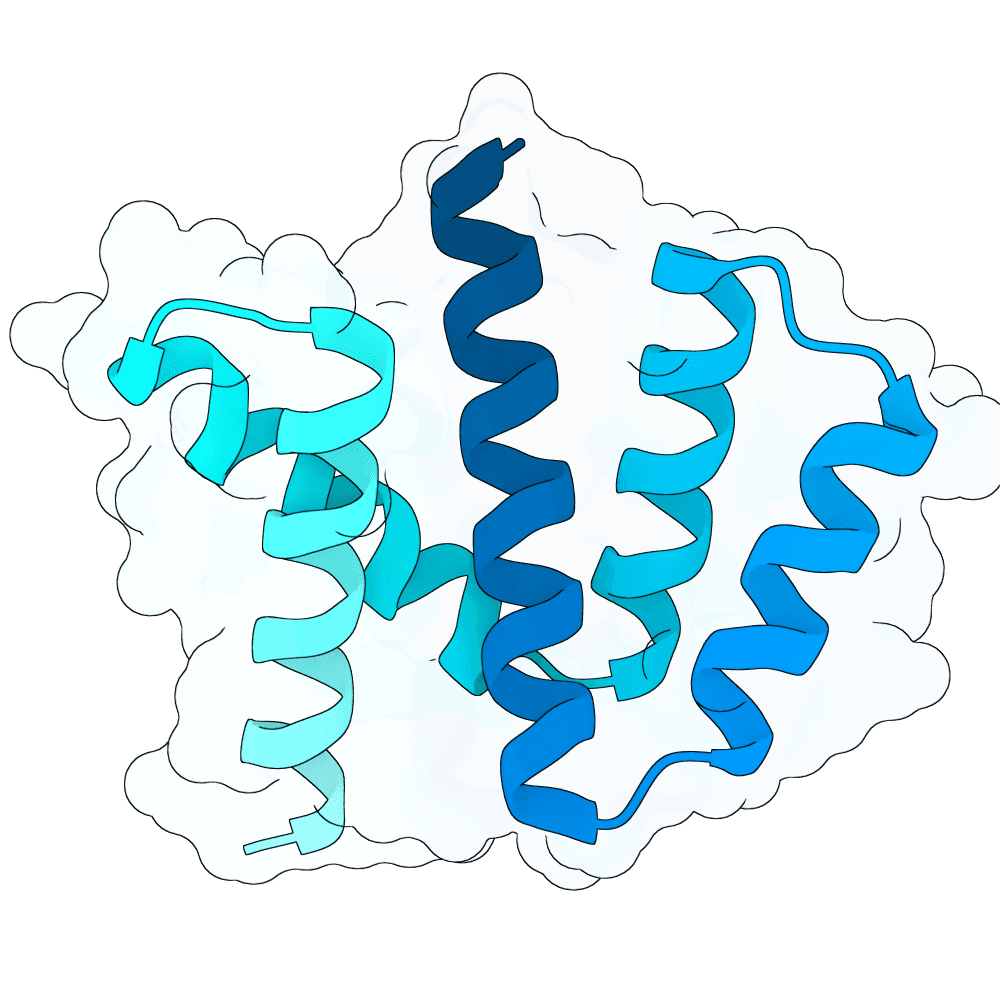
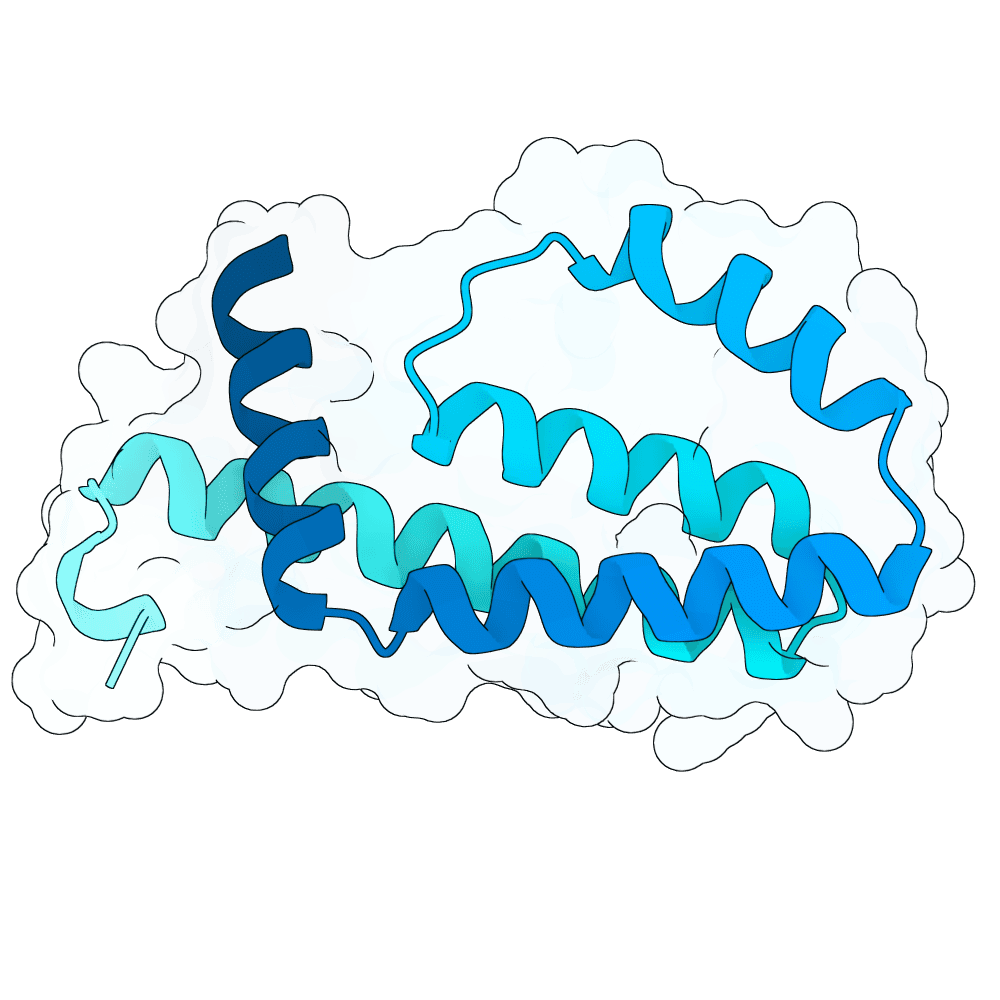
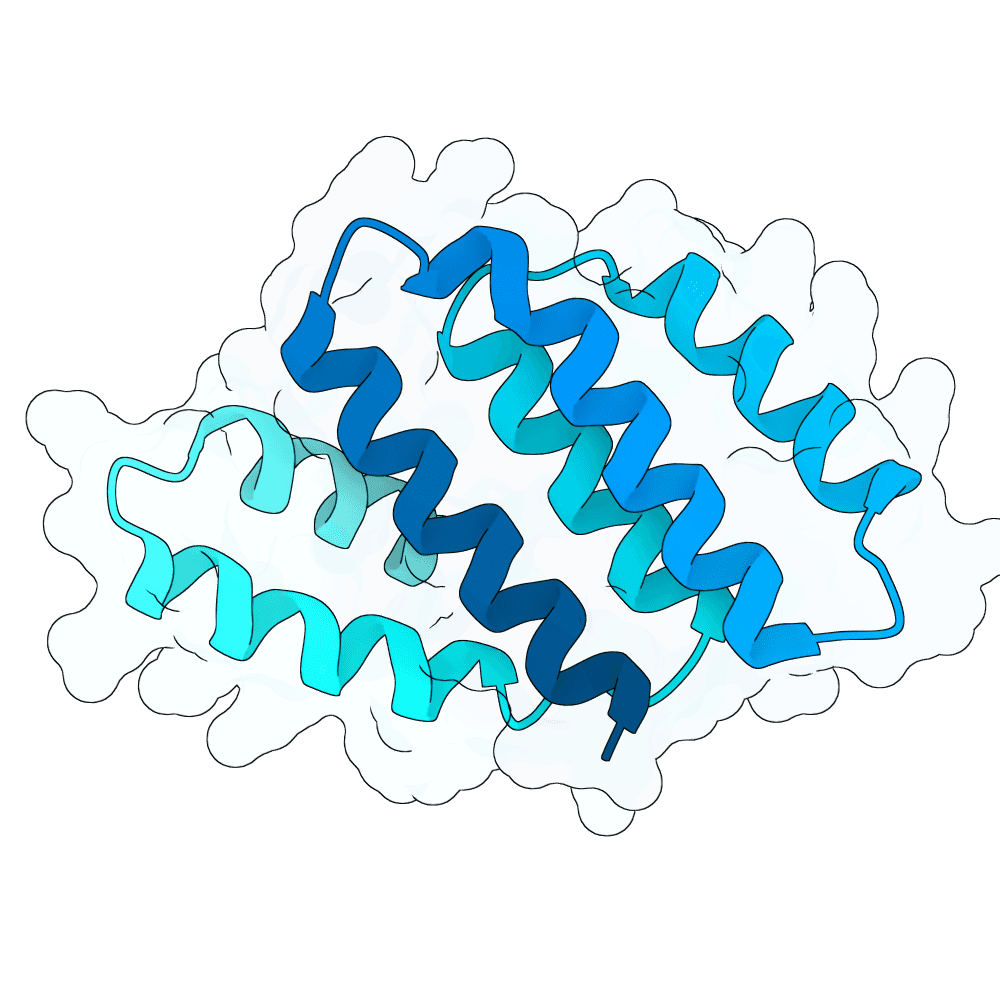
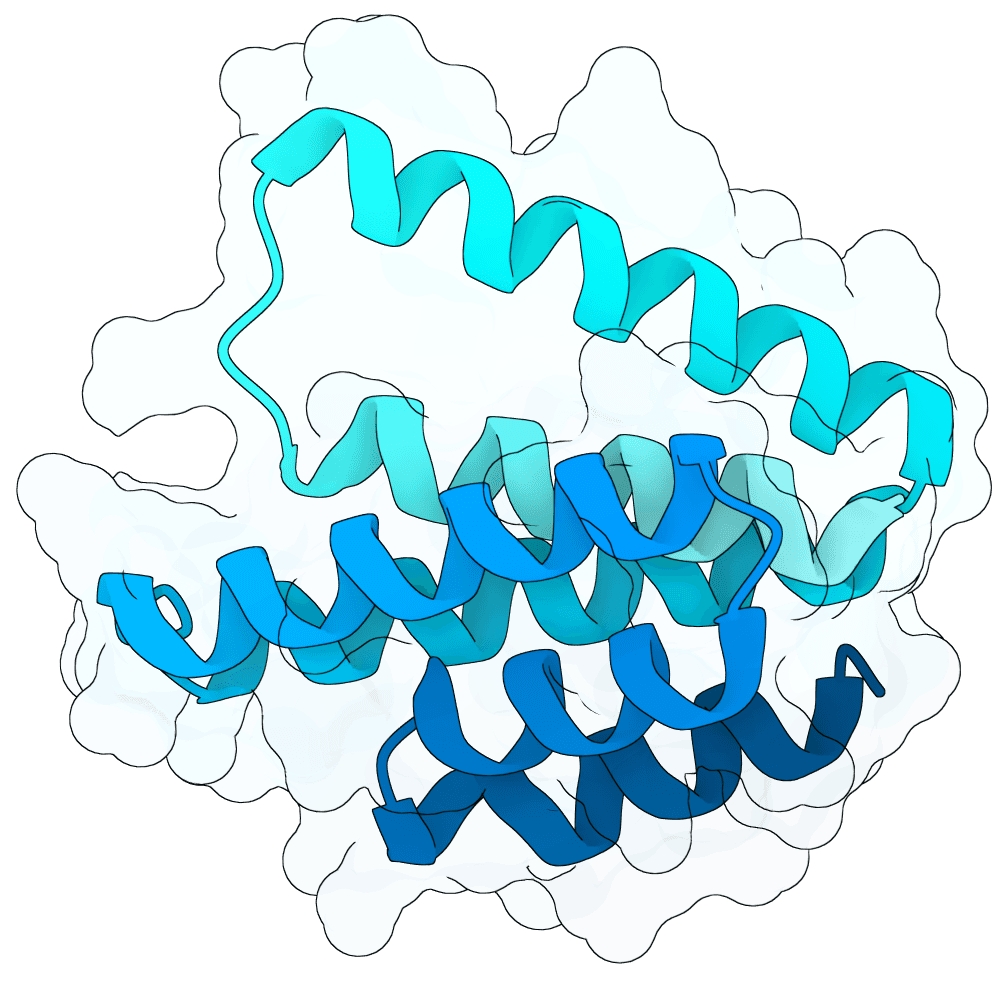
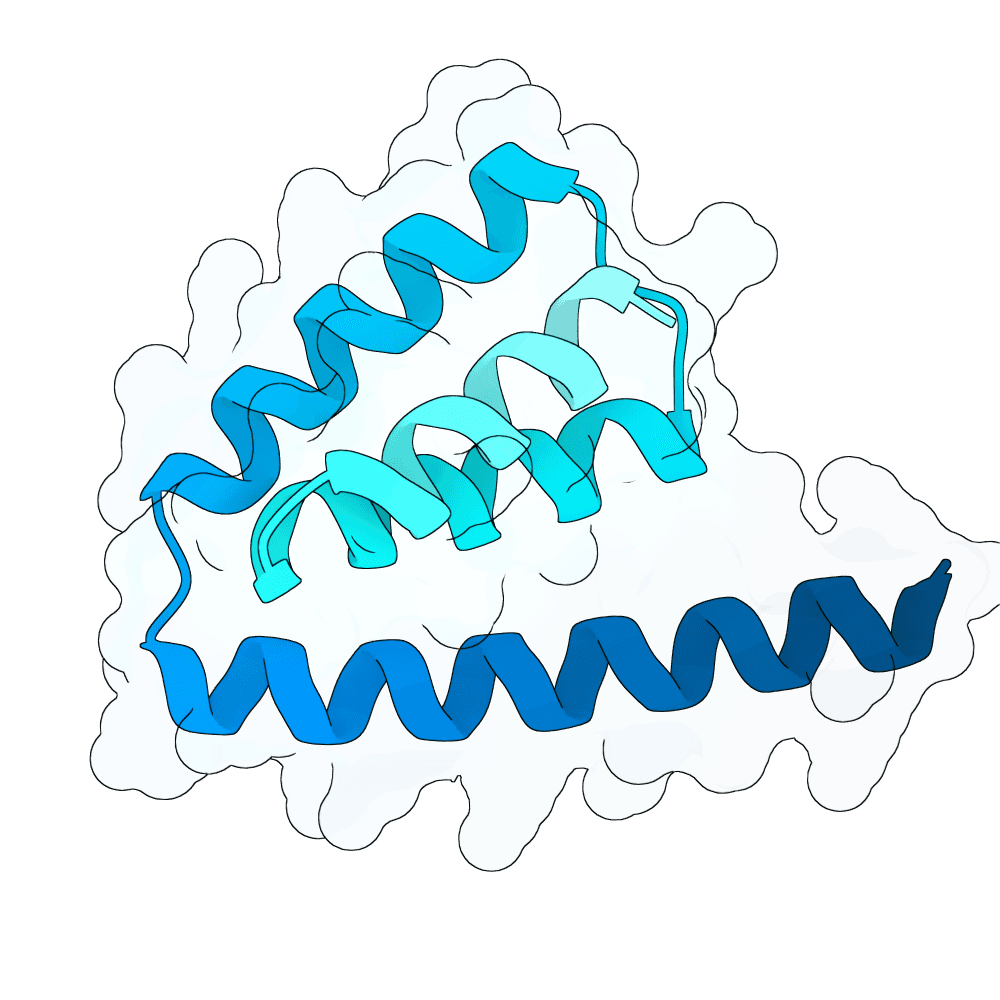
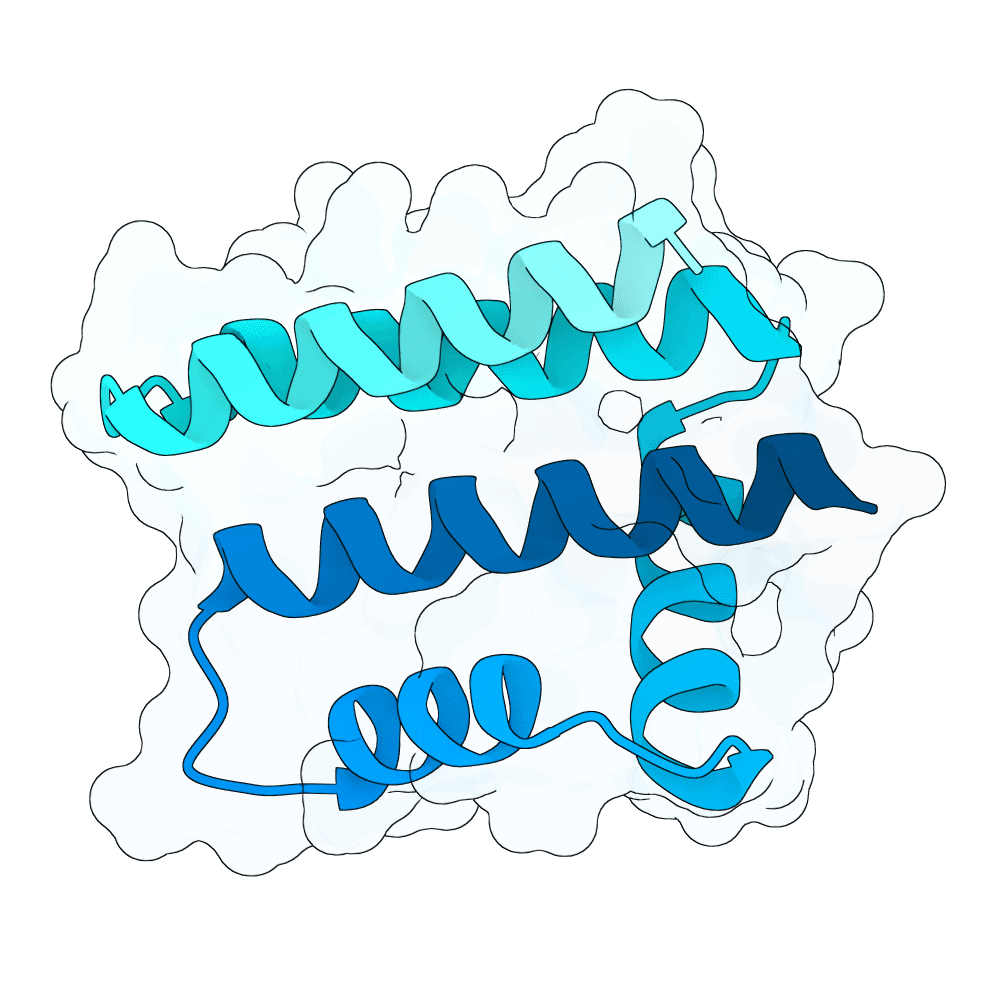
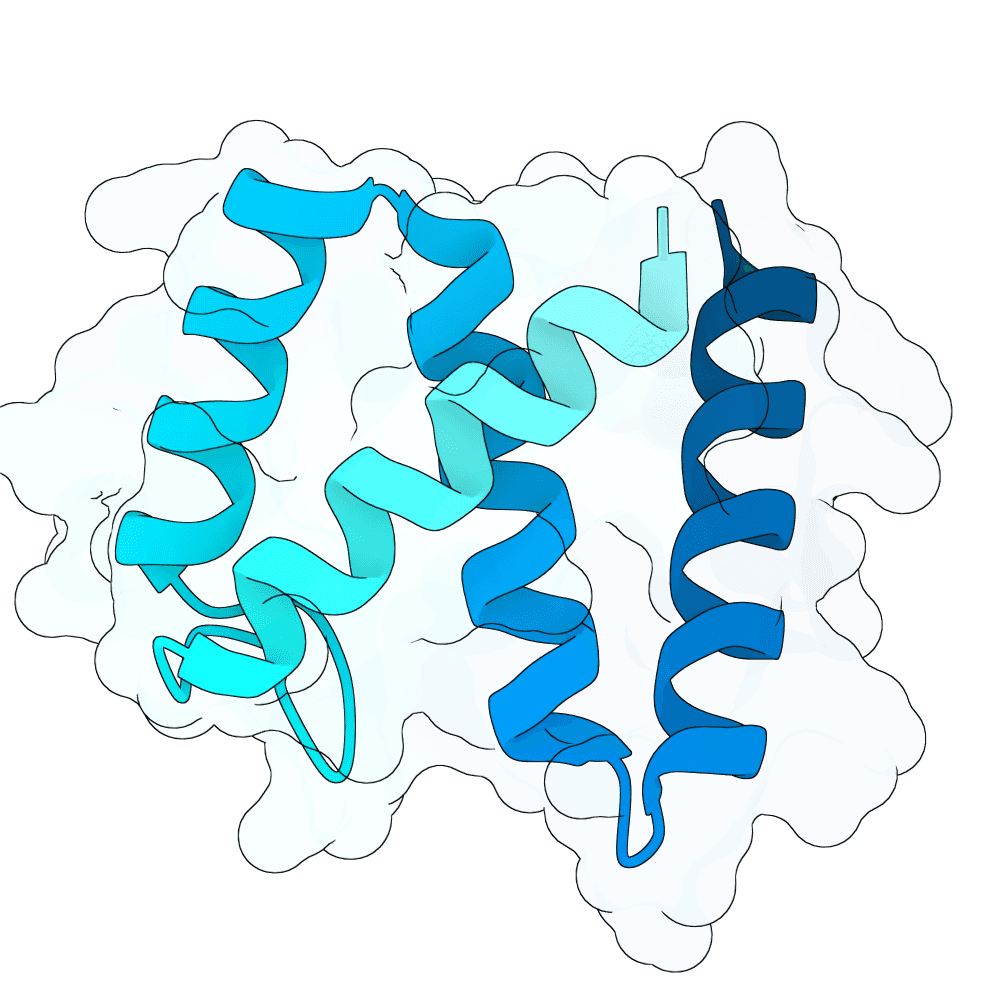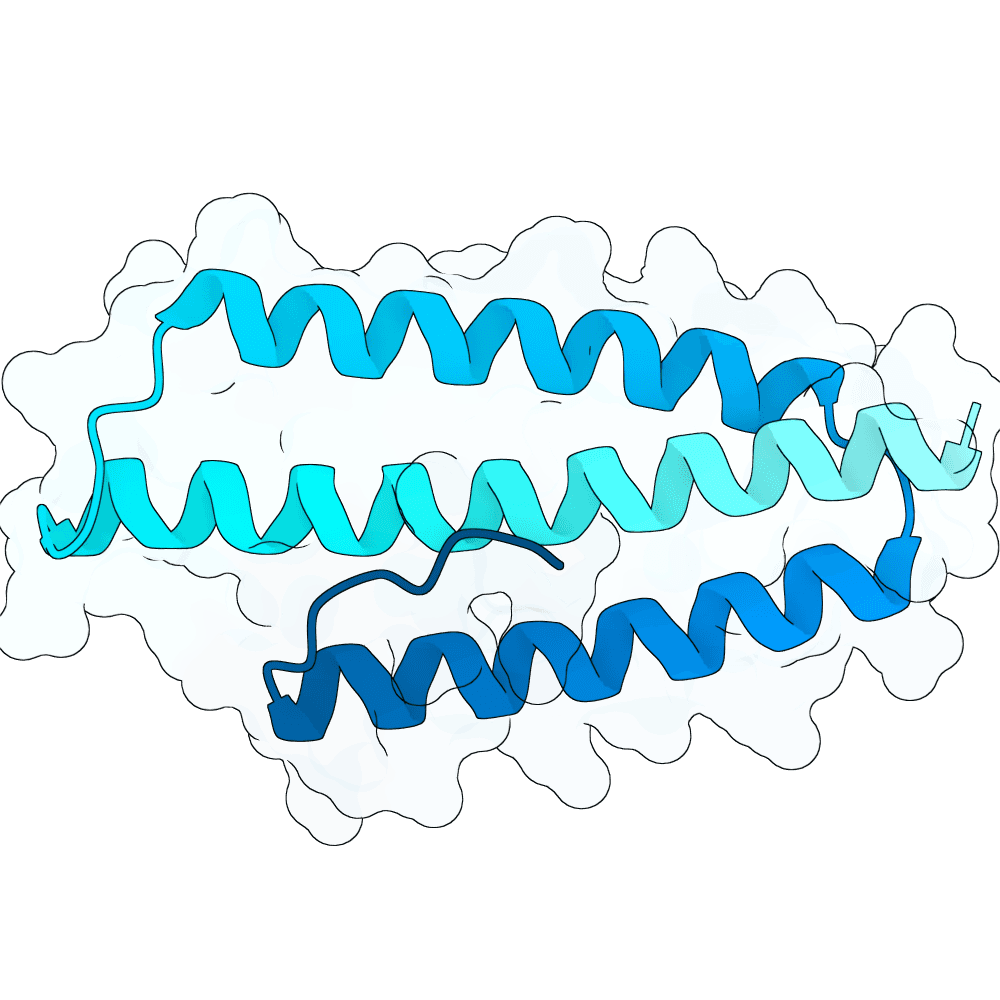Description
The designs are generated using de novo protein structure diffusion model that will be released soon. It extends Genie2 with atomized inputs, improved sampling, and optimized conditioning strategies. The model is trained on large-scale structure and interaction datasets with training tasks focused on binder design. Designs are filtered with more robust in-silico evaluation pipeline.