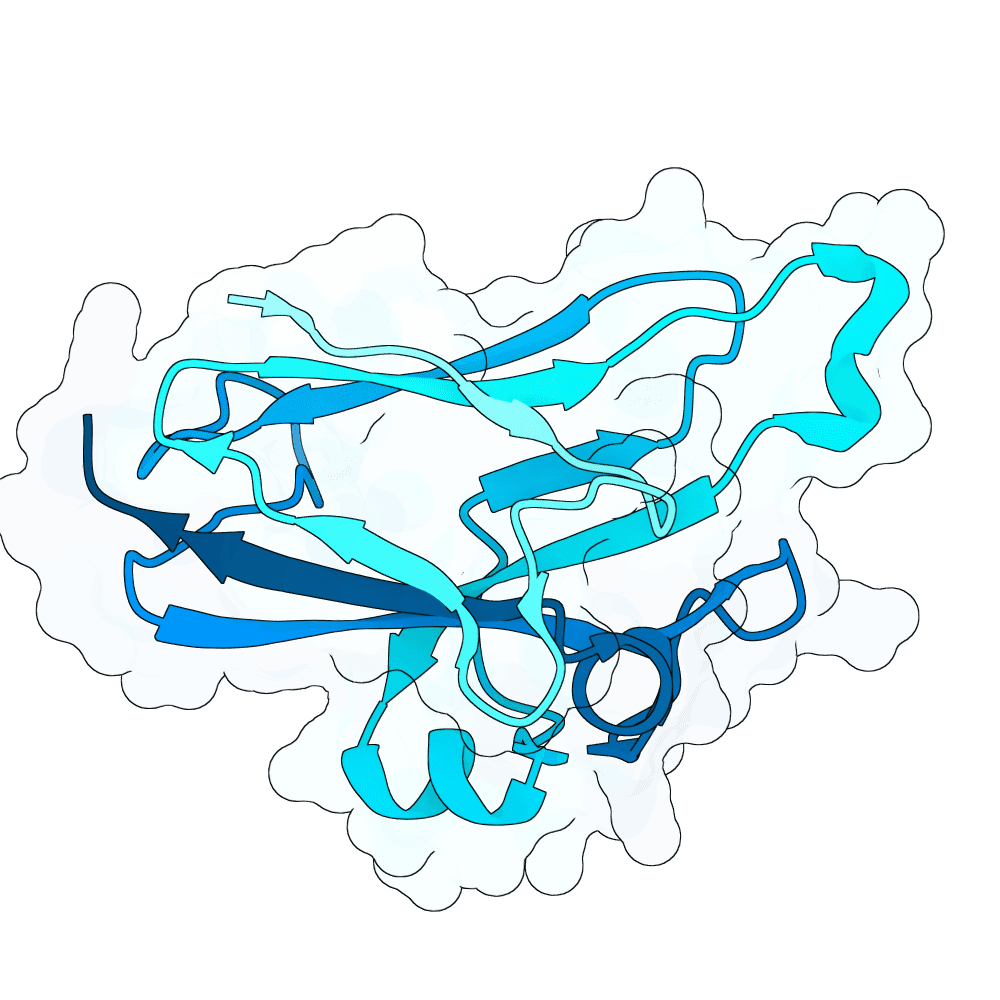
Description
Submission Overview
I used a multi objective lead optimization strategy to refine existing binder sequences, with continuous optimization in logit space around known leads.
Optimization procedure
- Start from binders with detectable activity and build target binder complexes with AlphaFold
- Generate a small panel of structure guided variants around these complexes and evaluate them with AlphaFold interface scores and Rosetta dG and SAP
- Embed all evaluated sequences with an ESM style protein language model and train a Gaussian process surrogate on these embeddings so that all objectives can be predicted directly from sequence [1]
- Run multi objective Bayesian optimization on top of this surrogate, using an expected hypervolume improvement style acquisition function as in BoTorch [2]
Sequence logit optimization and constraints
-
After each round, take the current Pareto optimal binders as leads
-
For each lead, initialize amino acid logits from its one hot sequence and optimize these logits with gradient based methods to maximize the acquisition value
-
During this optimization, apply two penalties
-
a penalty on the expected number of mutations relative to the parent, enforcing a target mutation radius for lead optimization
-
a penalty on divergence from the language model prior, keeping designs within a natural sequence distribution
-
The optimized logits define a local sequence distribution around each lead; from this distribution, sample discrete variants, rank them by acquisition value, and evaluate only the top candidates with AlphaFold and Rosetta
-
Add the new data to the training set, retrain the surrogate, update the Pareto set, and repeat for several generations
Choice of objectives and supporting work
- For AlphaFold based scoring I used ipSAE style interface metrics, which are now widely used as improved interface confidence measures over ipTM [3] and are supported by recent meta analyses on binder design success [4]
- Rosetta binding energies and packing terms were treated as complementary, more explicitly physical objectives, motivated by results on combining AlphaFold models with Rosetta style scoring for reliable complex assessment [6]
- The overall design of the workflow is consistent with modular multi objective binder pipelines such as BinderFlow [5]
Retrospective evaluation of final binders
- predict the complex with AlphaFold3 Server
- compute ipSAE using the official ipSAE script on the AF3 server models
- run Rosetta MinMover relax and then compute dG (REU) and SAP on the relaxed structures
For the AF3 Server models, the initial wild type and the main final designs scored as follows
- Initial_WT_AF3Server ipSAE 0.7035 dG −69.7 SAP 73.4
- pareto_1 ipSAE 0.7093 dG −81.6 SAP 70.4
- pareto_2 ipSAE 0.7744 dG −73.7 SAP 73.8
- pareto_3 ipSAE 0.7731 dG −65.9 SAP 68.7
- pareto_4 ipSAE 0.7686 dG −74.4 SAP 65.5
- pareto_5 ipSAE 0.7673 dG −73.6 SAP 74.8
- pareto_7 ipSAE 0.7630 dG −76.6 SAP 76.7
- pareto_8 ipSAE 0.7656 dG −74.0 SAP 77.0
- pareto_11 ipSAE 0.7782 dG −71.4 SAP 75.0
- pareto_12 ipSAE 0.7779 dG −71.7 SAP 74.6
- pareto_13 ipSAE 0.7813 dG −64.8 SAP 67.9
For comparison, the AF3 local baseline (Initial_WT_AF3Local) scored ipSAE 0.6997, dG −63.3, SAP 77.4. Across the final set, many designs simultaneously improve ipSAE and Rosetta dG relative to the starting binder, with SAP spanning a range that reflects the intended multi objective trade offs. This is consistent with the goal of pushing the Pareto front outward while staying within a controlled mutation distance of the original leads.
References
- [1] Lin Z et al. Evolutionary scale prediction of atomic level protein structure with a language model. Science 2023.
- [2] Balandat M et al. BoTorch a framework for efficient Monte Carlo Bayesian optimization. NeurIPS 2020.
- [3] Dunbrack RL Jr. Res ipSAE loquunt what is wrong with AlphaFold ipTM and how to fix it. bioRxiv 2025.
- [4] Overath MD et al. Predicting experimental success in de novo binder design a meta analysis of 3,766 experimentally characterised binders. bioRxiv 2025.
- [5] González Rodríguez N et al. Automated and modular protein binder design with BinderFlow. PLOS Computational Biology 2025.
- [6] Harmalkar A et al. Reliable protein protein docking with AlphaFold Rosetta and cryo EM. eLife 2025.