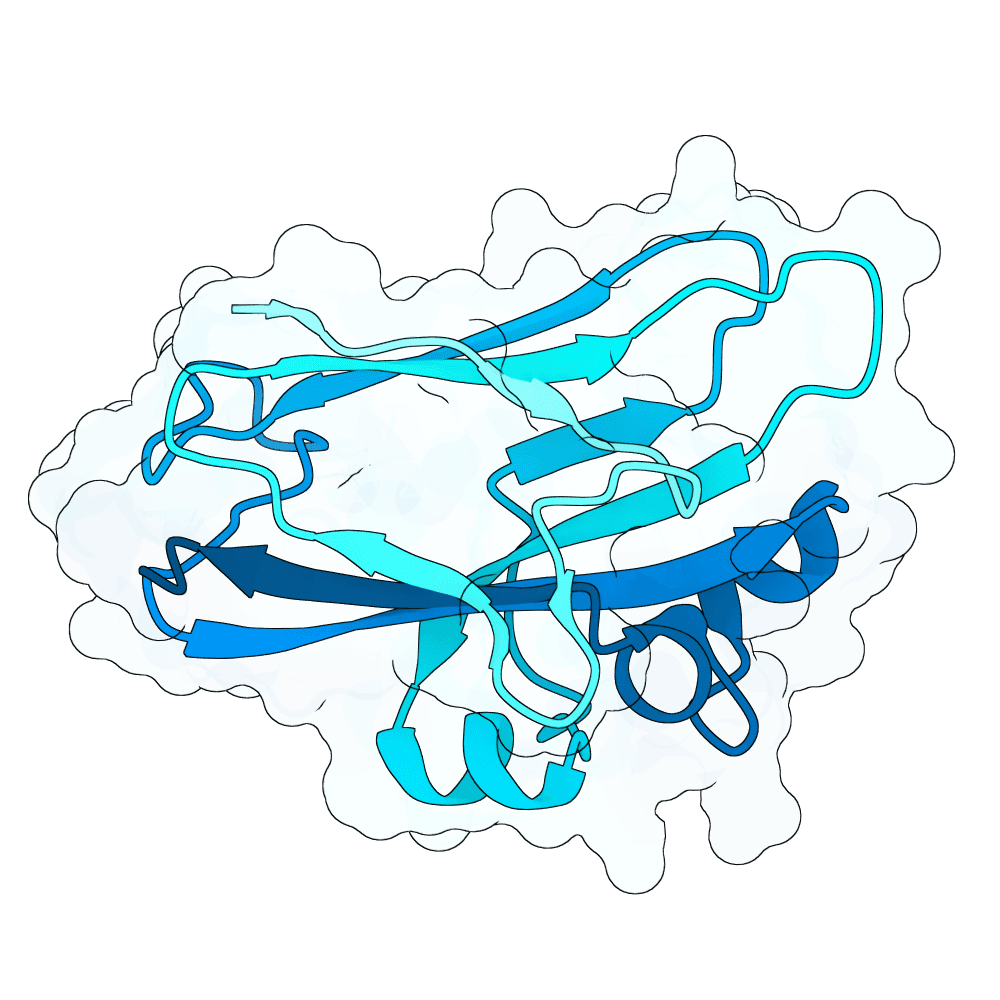
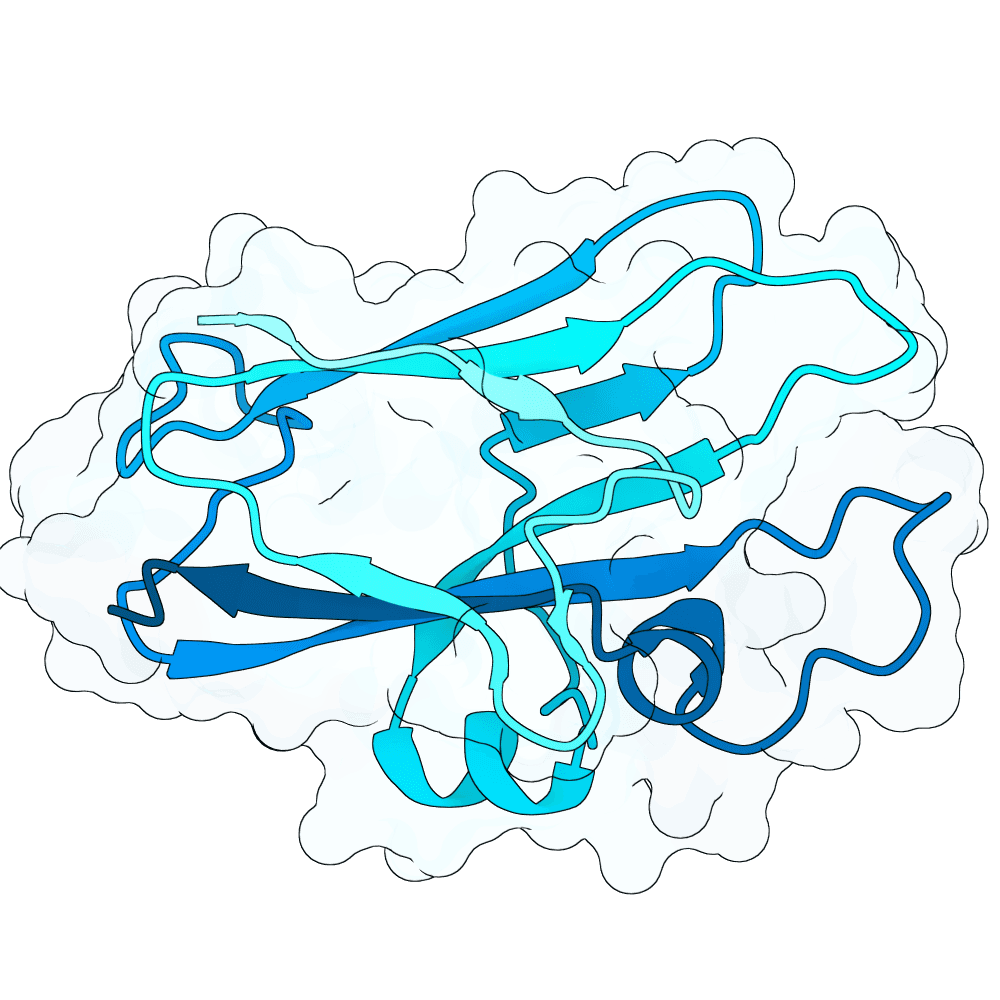
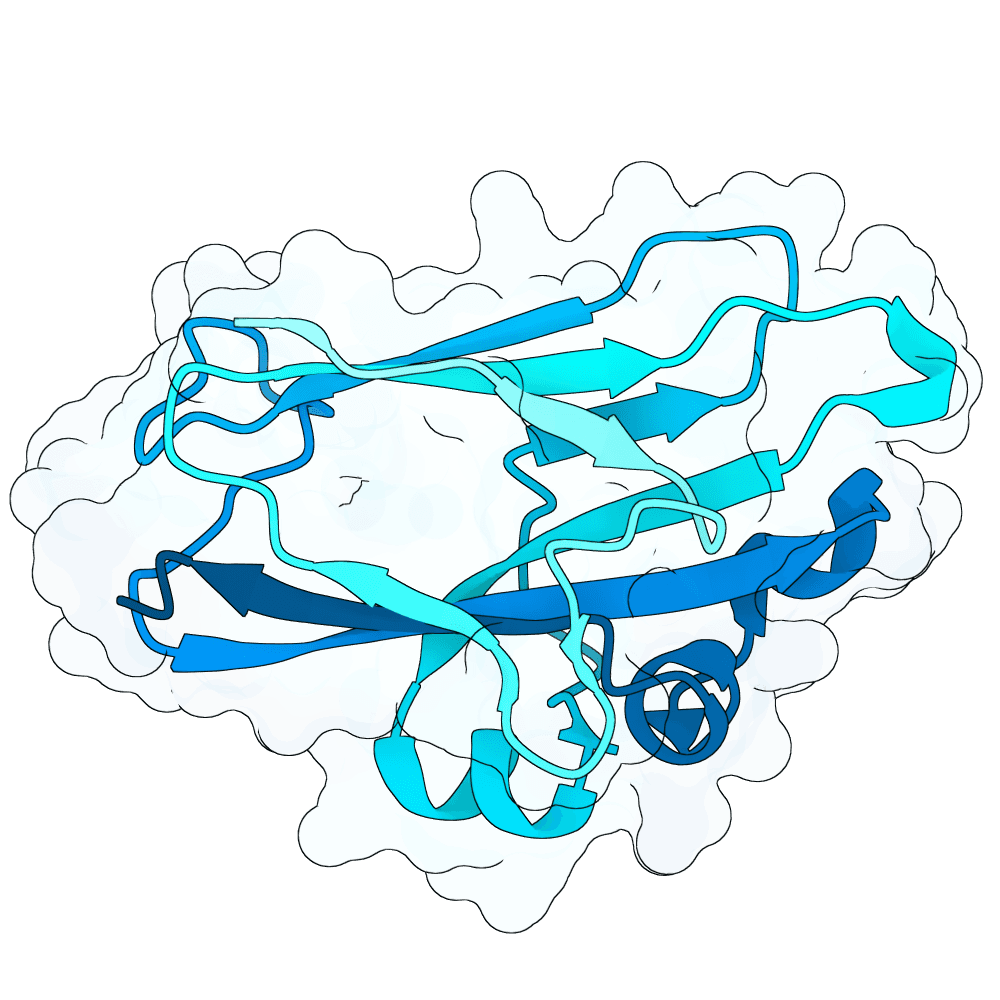
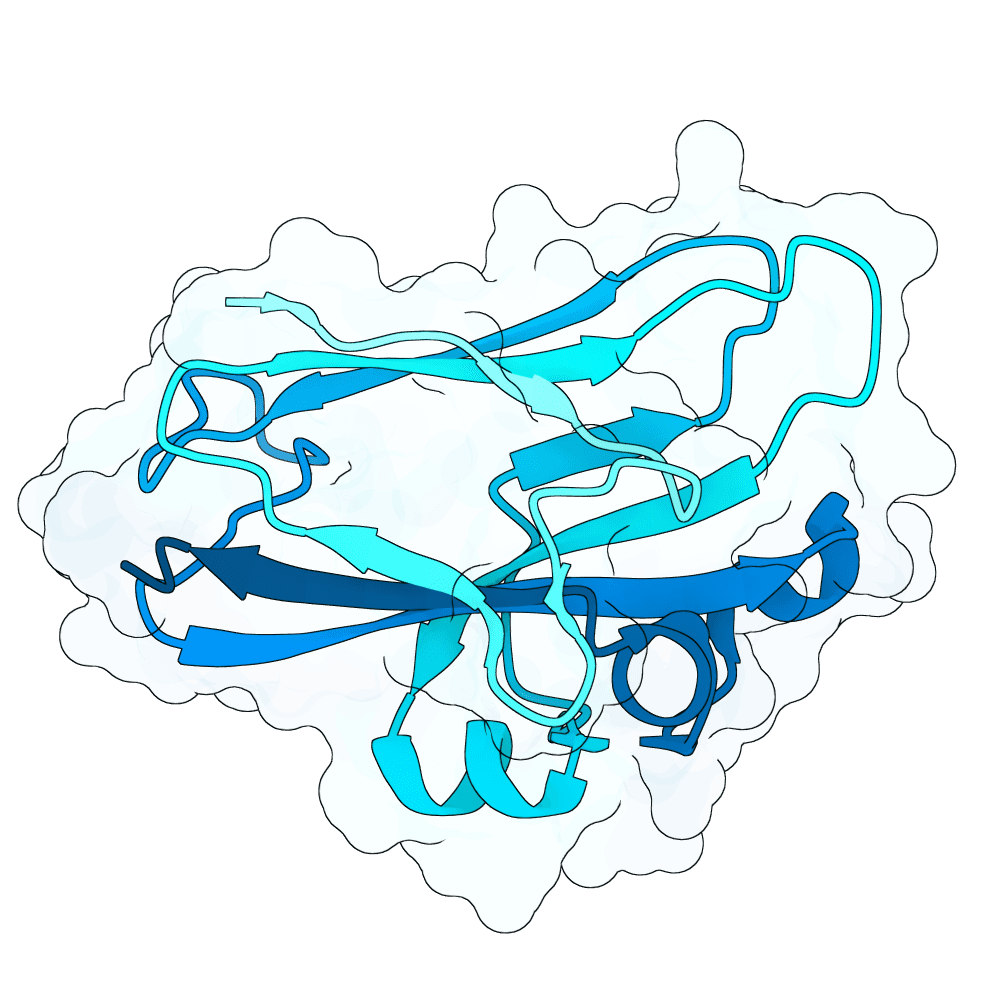
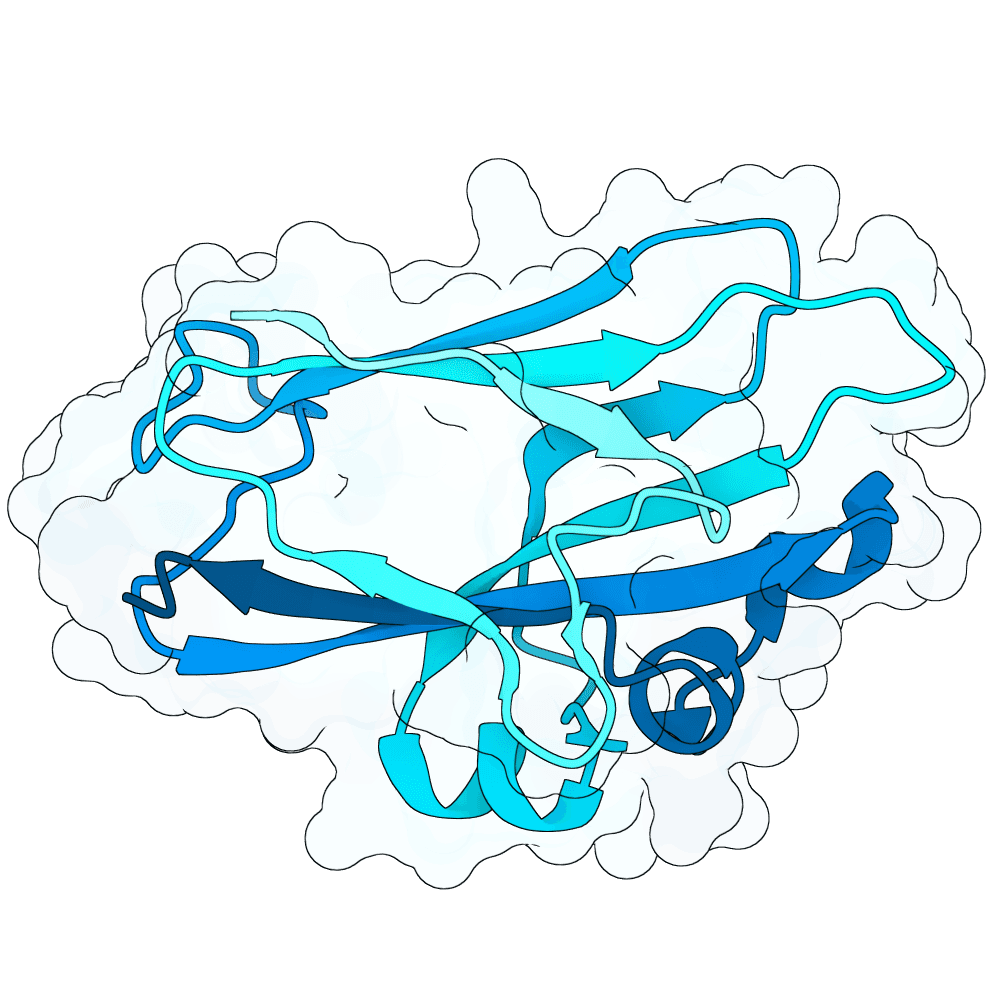
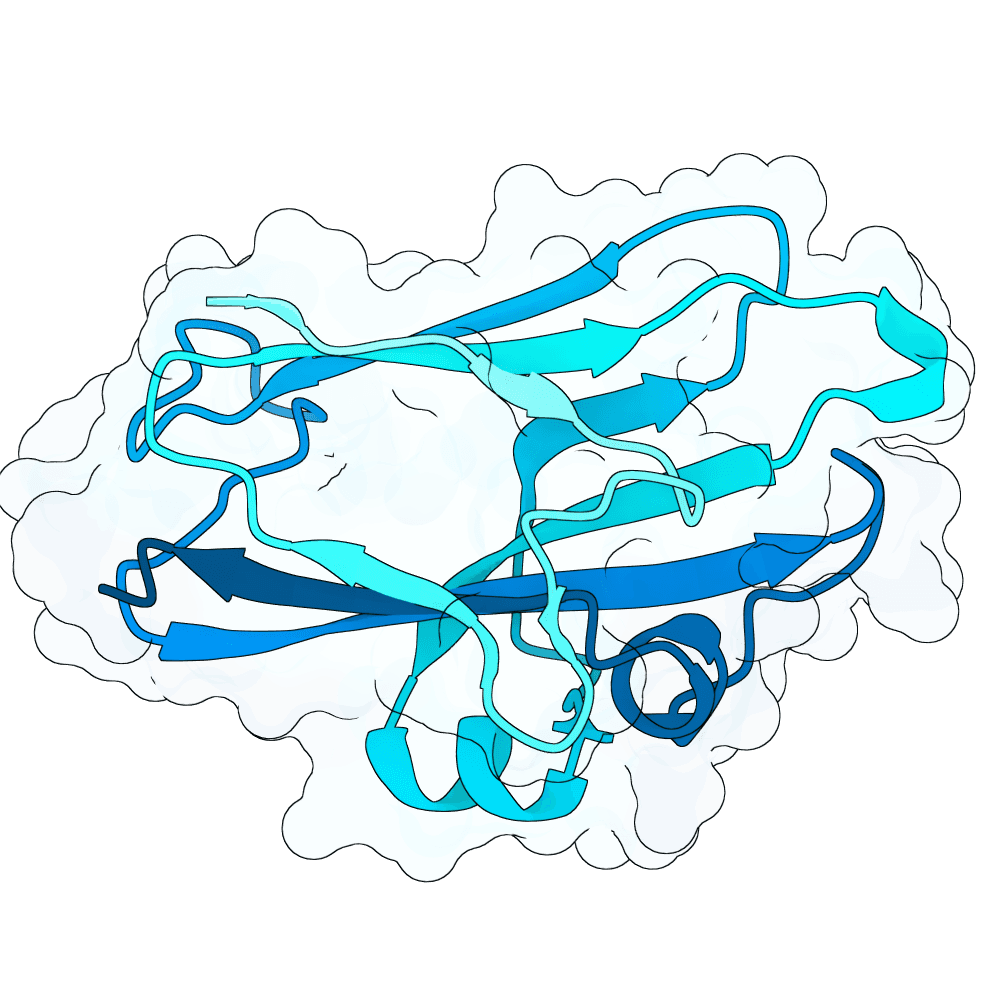
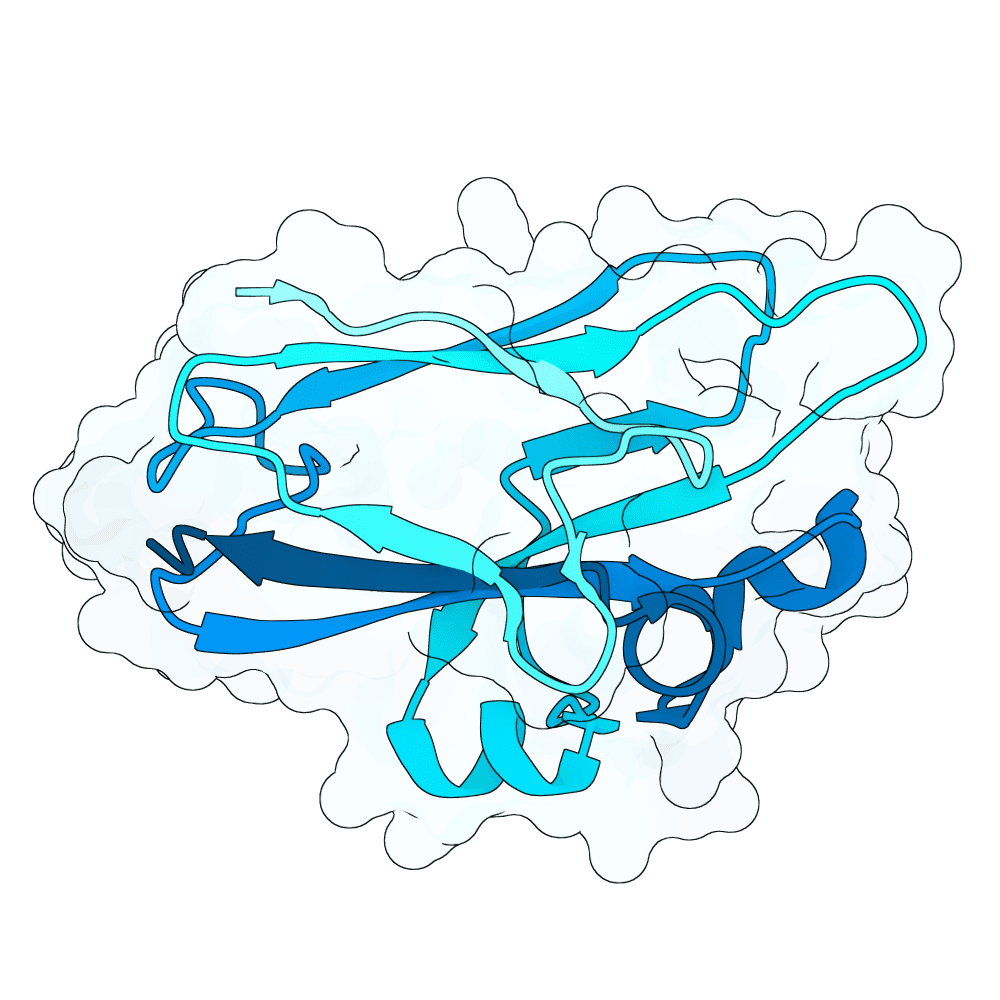
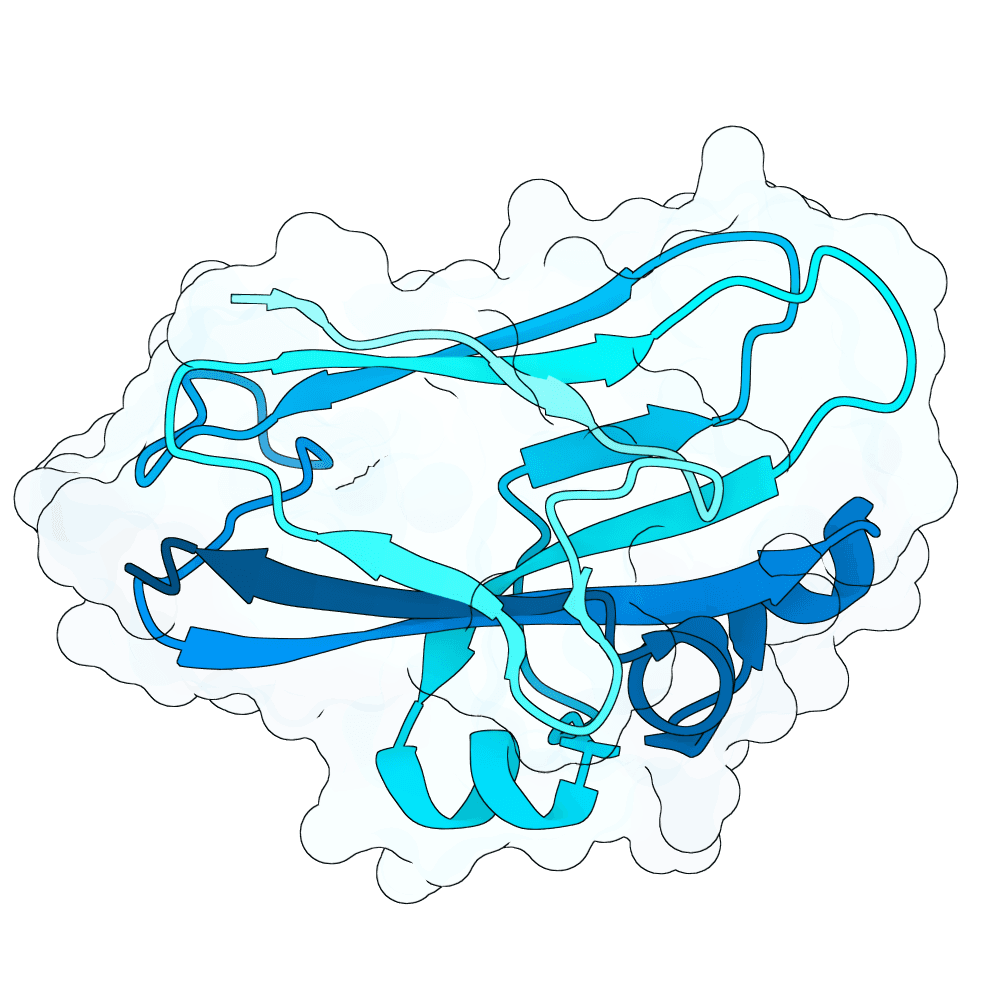
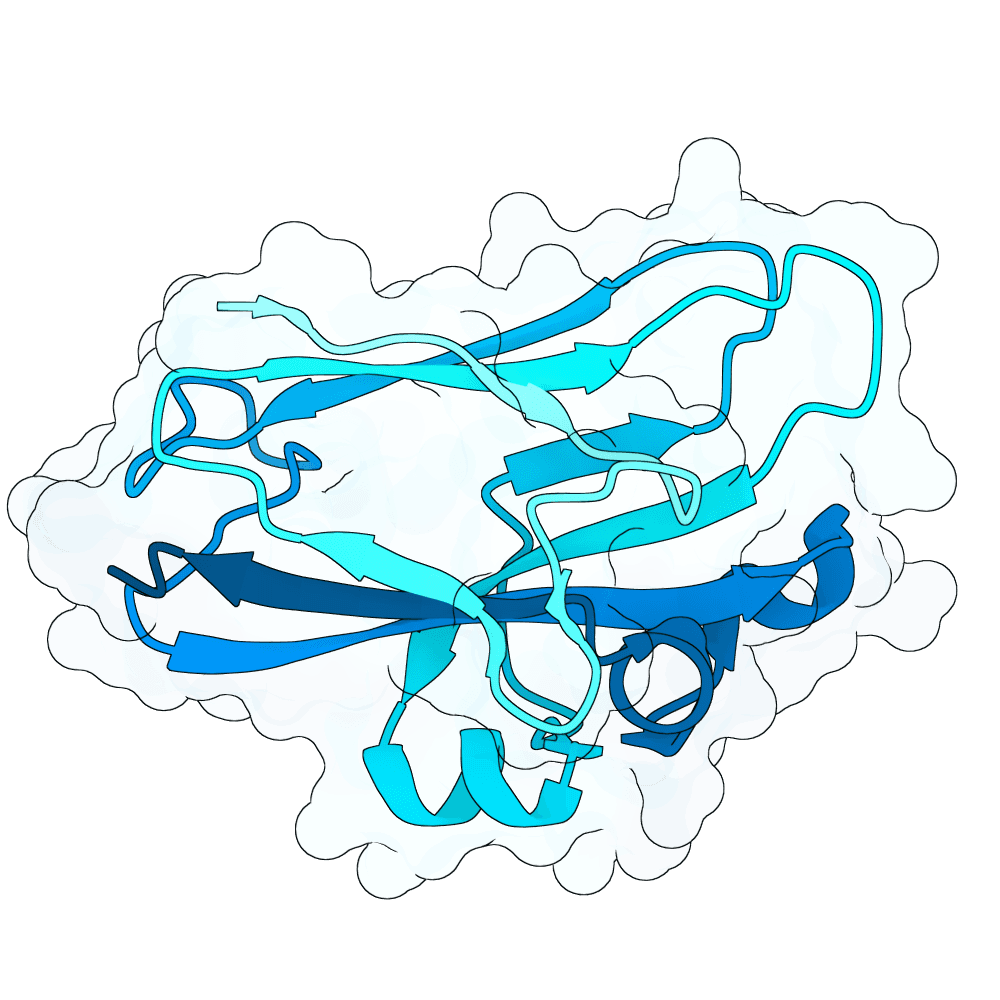
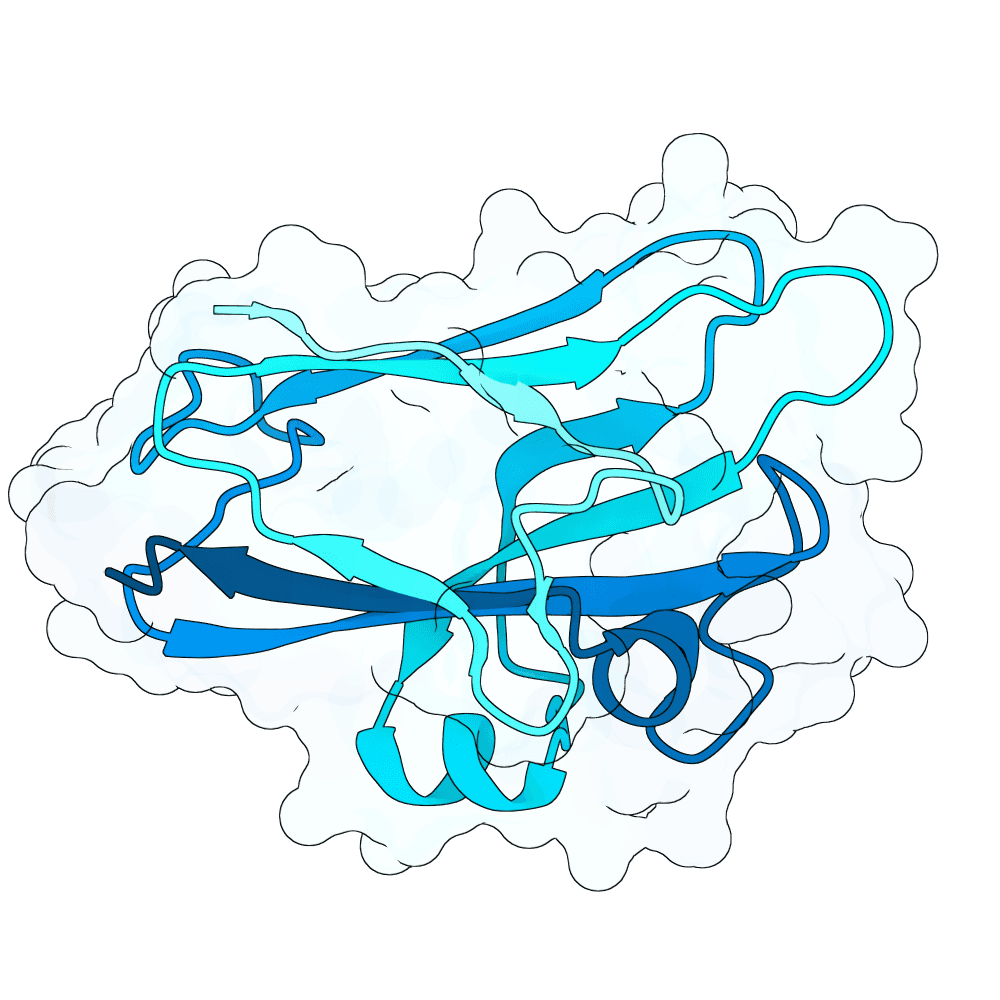
Description
As a part of my master's project, I created a simple nanobody design pipeline consisting of: RFDiffusion for backbone design -> ProteinMPNN for sequence design -> Boltz-2 for structure design -> Rosetta Fast Relax -> ipSAE scoring and ranking. In this case, I created 1000 different backbones with 2 ProteinMPNN sequences each, leading to a total of almost 2000 designs. The design's originated from the 2vsm complex with mutated CDRs. Of the resulting nanobodies, I then chose the ones with the highest min_ipSAE scores obtained after a Boltz-2 structure design. The pipelines hyperparameters were tuned earlier when working with other targets (especially BCMA) in a way, to create a maximum metric discrimination between binders and non-binders. The binder design pipeline was run on an NVIDIA GeForce RTX 4090 GPU.