Description
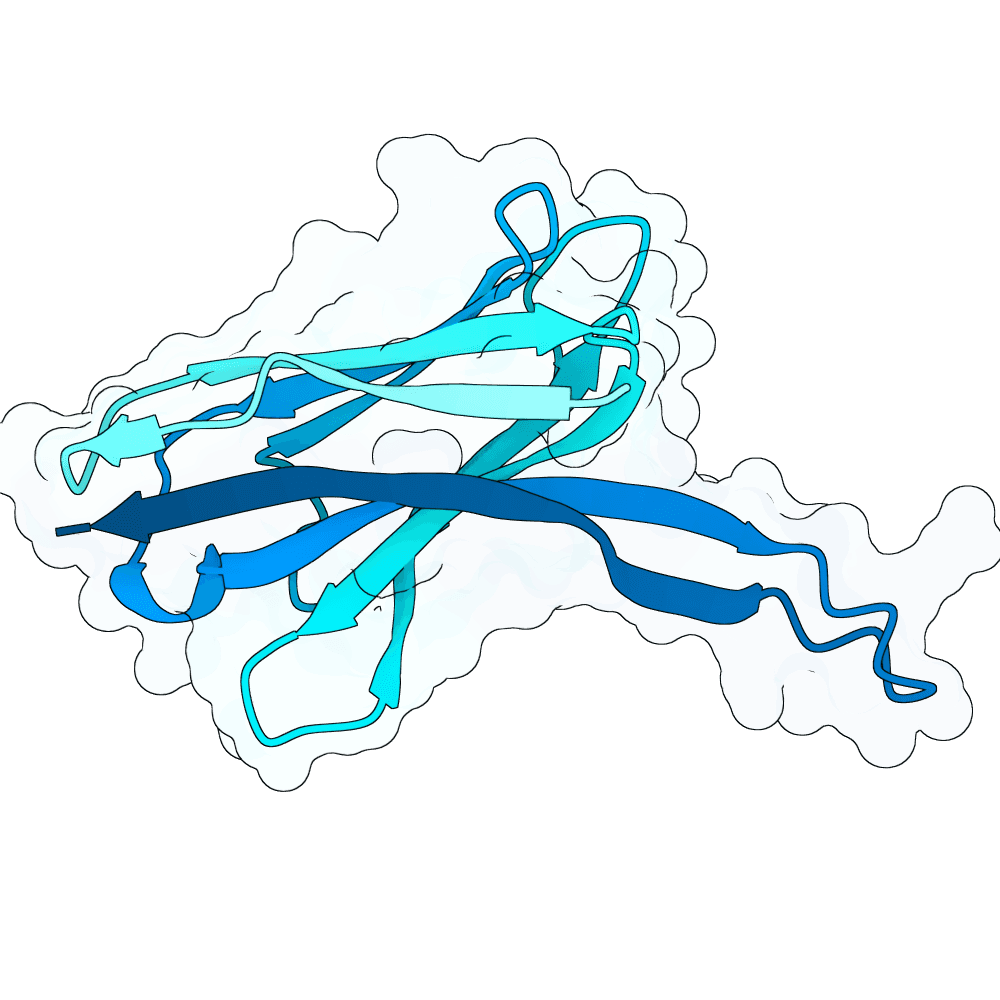
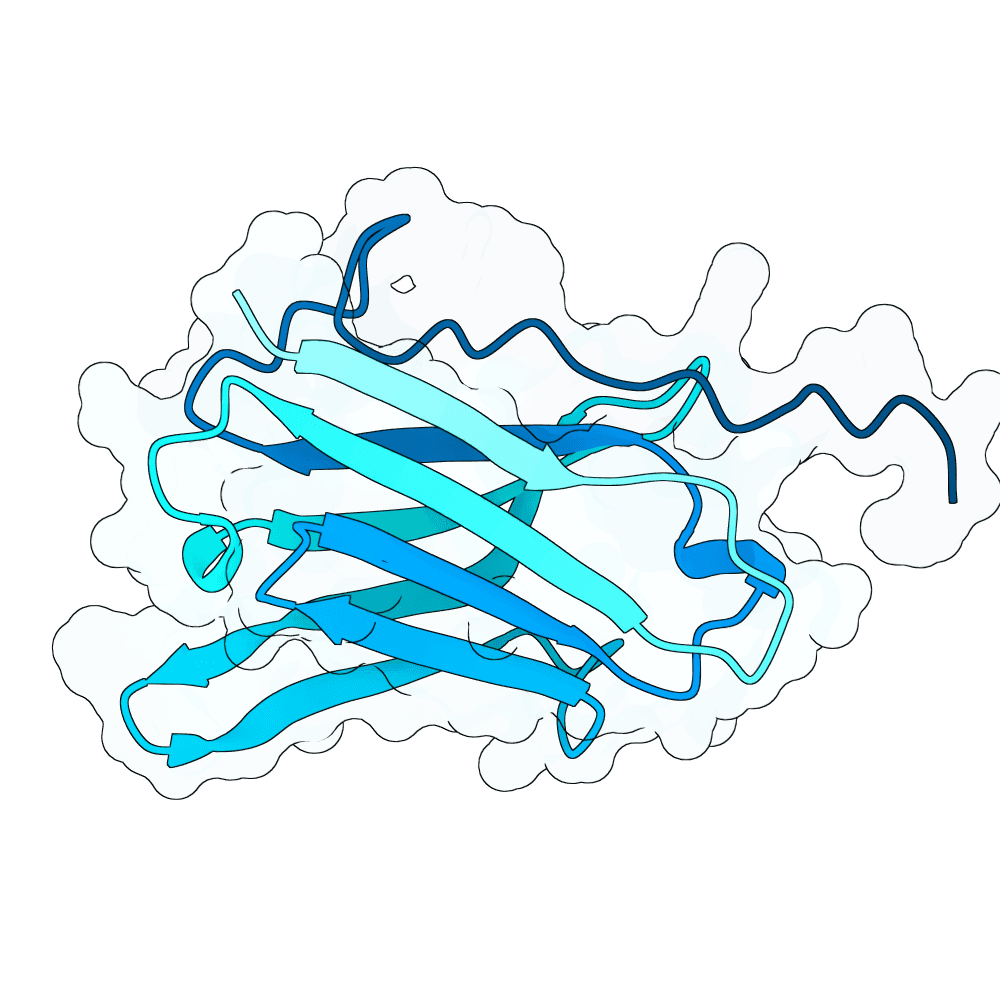
Methods: Agentic Structural Design and RL-Guided Protein Language Models Work by Romain Pastre and collaborators Center for Genomic Regulation (CRG), Ferruz Lab Method I: Agent-Guided Iterative Structural Design. -- We developed an autonomous feedback loop governed by a decision-making agent to navigate the design space iteratively. Each design cycle begins with backbone generation using RFdiffusion, conditioned on specific hotspots and Complementarity-Determining Region (CDR) lengths. This is followed by sequence recovery targeting specific binder regions using both ProteinMPNN and SolubleMPNN.The generated candidates undergo rigorous validation using Ablang2 for perplexity assessment and a comprehensive multiparametric scoring function. This scoring function evaluates candidates based on iPAE, pDockQ, dSASA, DeltaG per residue, unsaturated hydrogen bonds, and shape complementarity (sc) .To optimize local substructures, the top three candidates from each batch are subjected to an inverse folding refinement step via ProteinMPNN, which modifies specific residue positions based on their individual per-residue performance in the cited metrics. At the conclusion of each generation cycle, a comprehensive report is synthesized and fed back to a Llama 3 agent (specifically optimized for bioengineering tasks). The agent reasons over these results to algorithmically adjust hyperparameters for the subsequent design iteration. Method II: Supervised Fine-Tuning and Reinforcement LearningLeveraging the high-quality synthetic data generated in Method I -- we advanced to a sequence-based approach using a large proprietary Protein Language Model (pLM) developed at the Ferruz Lab.Supervised Fine-Tuning (SFT):We first performed supervised fine-tuning on the pLM using the curated sequences obtained from the agent-guided structural design loop. This stage adapted the model's sampling distribution to reflect the structural and functional motifs identified in the previous method.Reinforcement Learning (RL): To further align the model with functional requirements, we applied Reinforcement Learning to the SFT version of the model. We utilized a reward function derived from the key structural metrics established in Method I (including iPAE and pDockQ). This process directed the model to prioritize the generation of sequences that maximize these stability and binding affinity indicators.