Description
The tool we used to design these binders is Prometheus, a protein design method that uses an inverse folding model as a reinforcement learning agent to achieve self-optimization.
The tool we used to design these binders is Prometheus, a protein design method that uses an inverse folding model as a reinforcement learning agent to achieve self-optimization.
id: vast-quail-sand
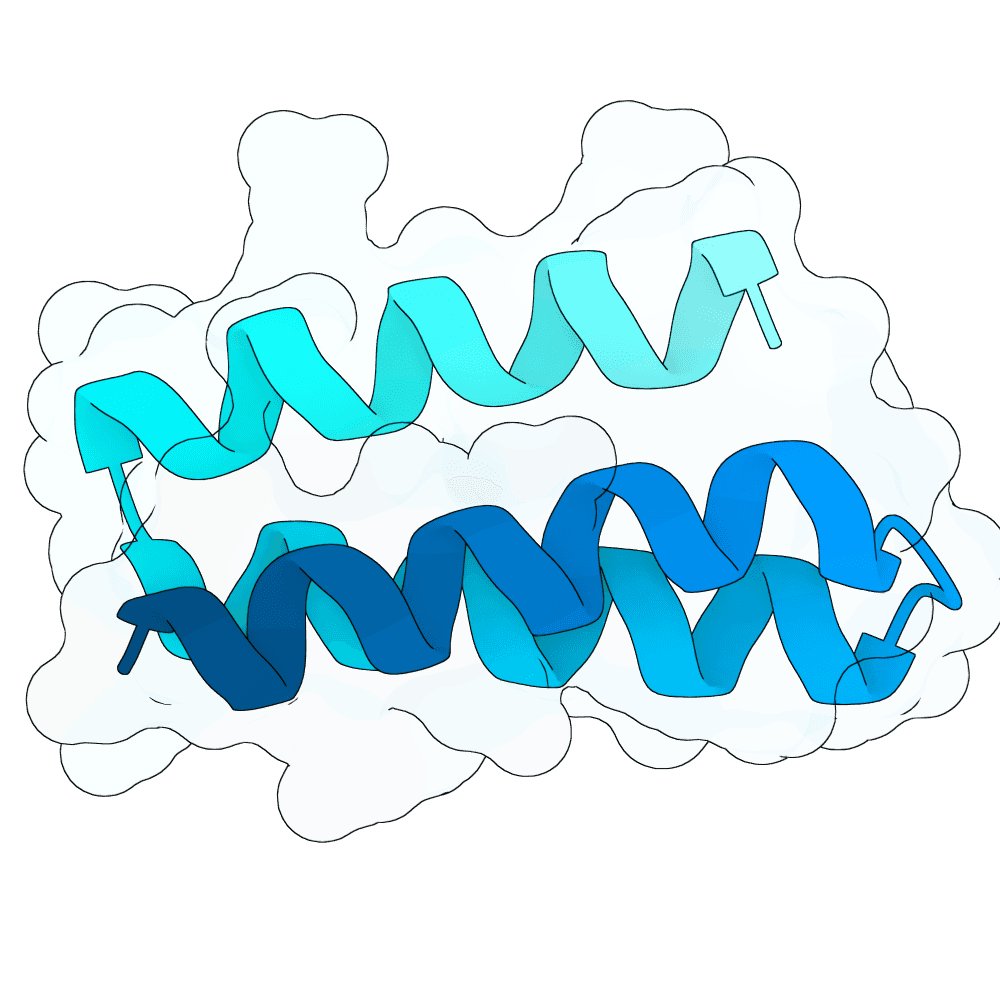
Nipah Virus Glycoprotein G
None
89.30
True
5.8 kDa
56
id: quick-deer-snow
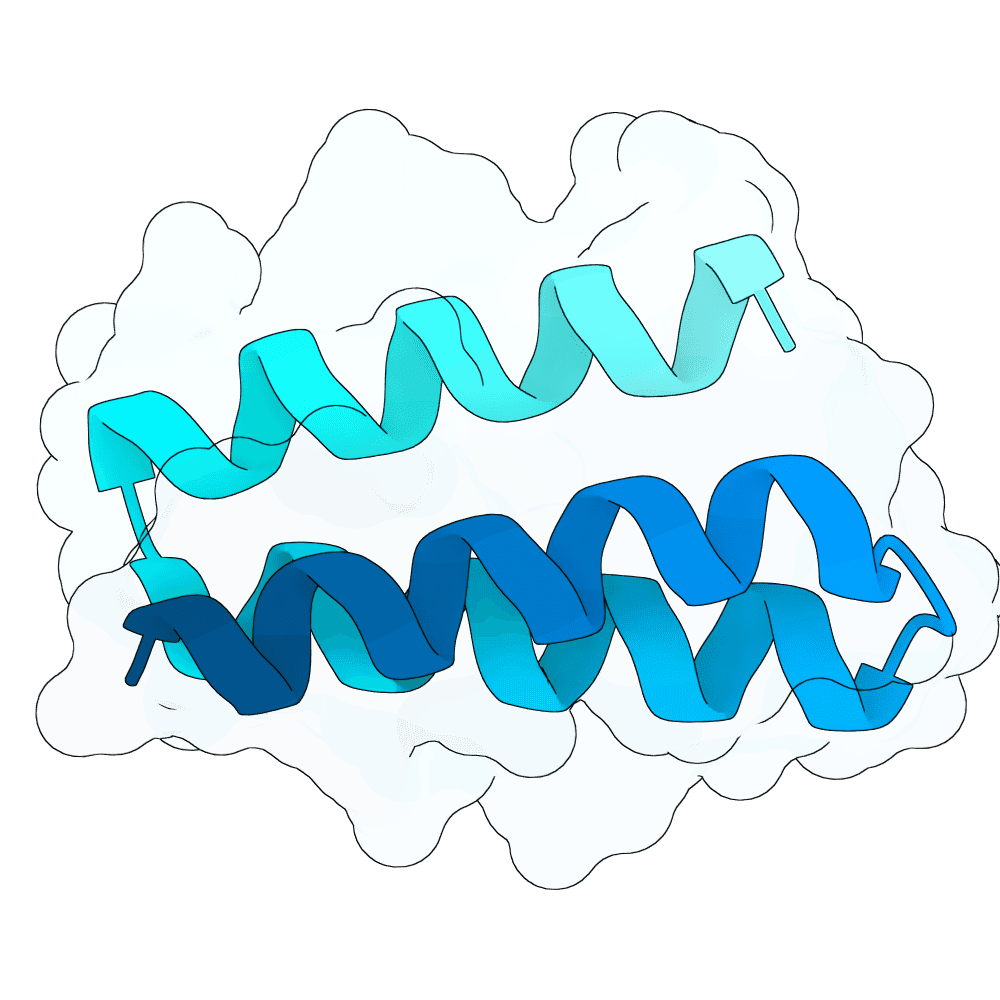
Nipah Virus Glycoprotein G
None
87.64
True
6.1 kDa
56
id: jade-cobra-thorn
Nipah Virus Glycoprotein G
None
88.23
False
6.1 kDa
56
id: soft-panda-rose
Nipah Virus Glycoprotein G
None
89.94
False
5.9 kDa
56
id: ivory-deer-clay

Nipah Virus Glycoprotein G
None
91.61
True
5.5 kDa
56
id: golden-owl-onyx
Nipah Virus Glycoprotein G
None
88.55
True
5.9 kDa
56
id: quiet-panther-wave
Nipah Virus Glycoprotein G
None
88.14
True
6.1 kDa
56
id: silent-shark-rose

Nipah Virus Glycoprotein G
0.67
89.19
--
5.8 kDa
56
id: crimson-panther-plume
Nipah Virus Glycoprotein G
0.34
87.72
--
6.0 kDa
56
id: amber-toad-willow
Nipah Virus Glycoprotein G
0.07
87.27
--
6.2 kDa
56