Proteins (4)
SSquirtle
id: green-orca-pearl
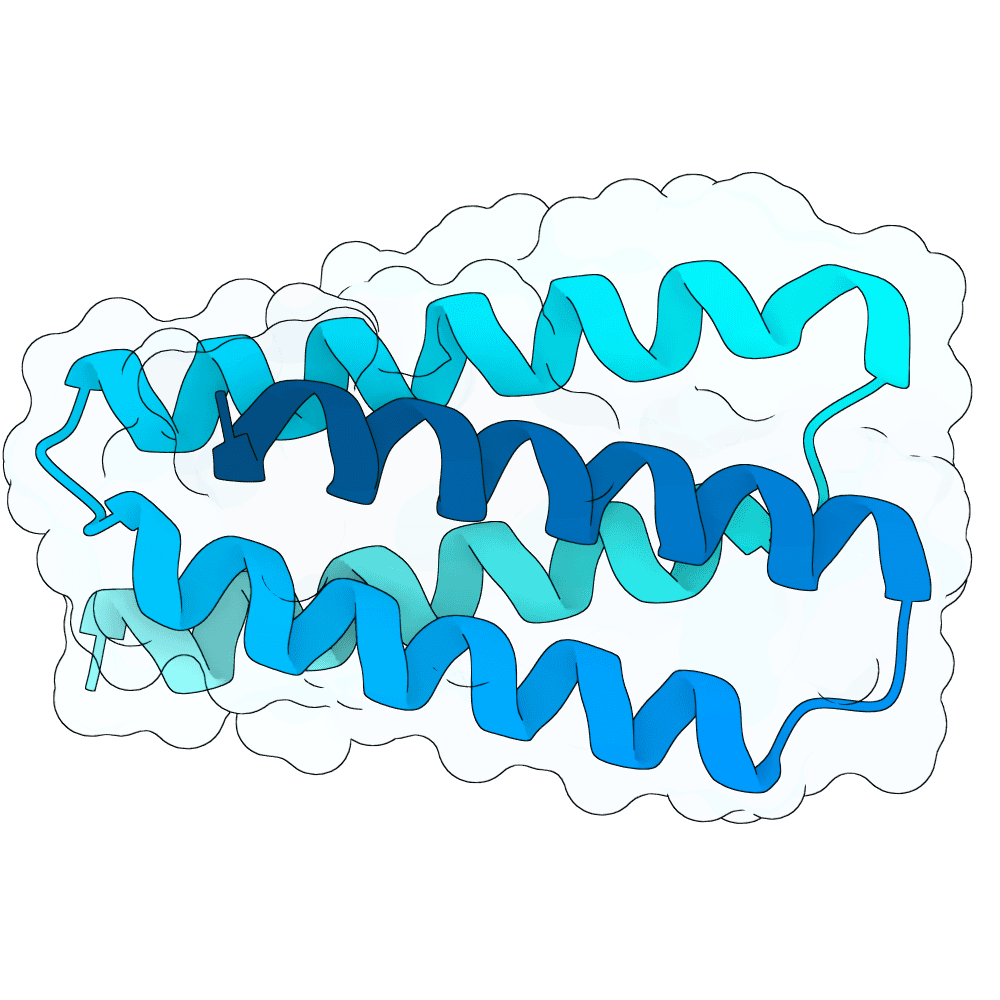
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
93.46
Expressed
False
Mol. Weight
10.7 kDa
AA Length
100
SSquirtle
id: crimson-owl-cedar
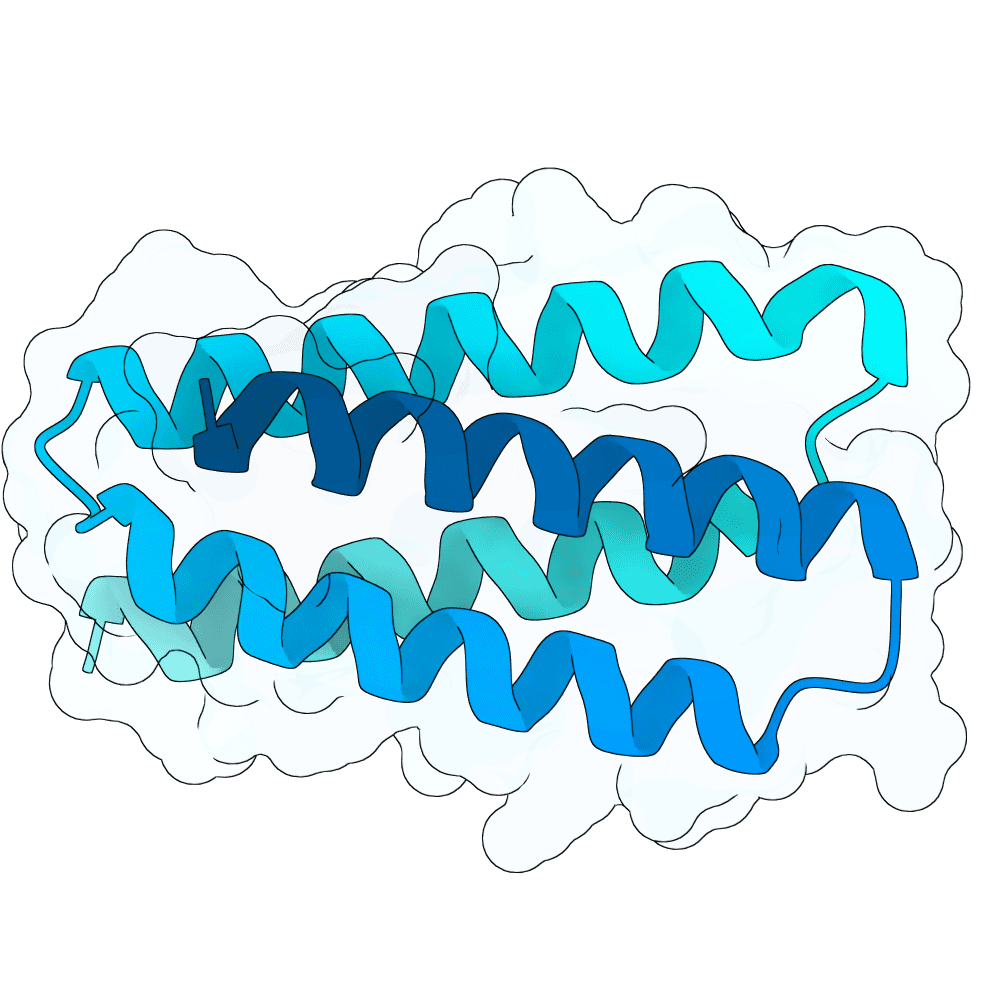
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
89.98
Expressed
True
Mol. Weight
11.6 kDa
AA Length
100
SSquirtle
id: golden-owl-willow
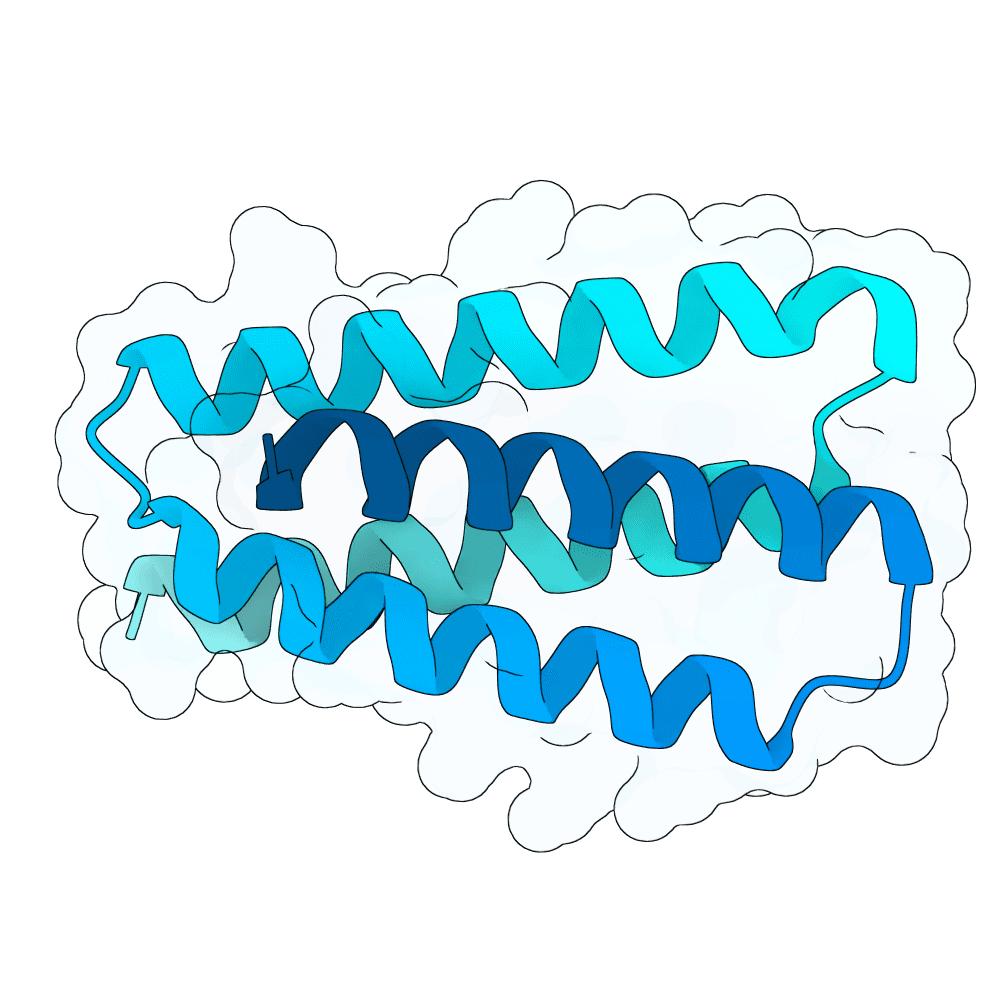
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
91.90
Expressed
True
Mol. Weight
11.0 kDa
AA Length
100
SSquirtle
id: gentle-bear-reed
Target
Nipah Virus Glycoprotein G
Binding
None
pLDDT
92.21
Expressed
True
Mol. Weight
11.1 kDa
AA Length
100