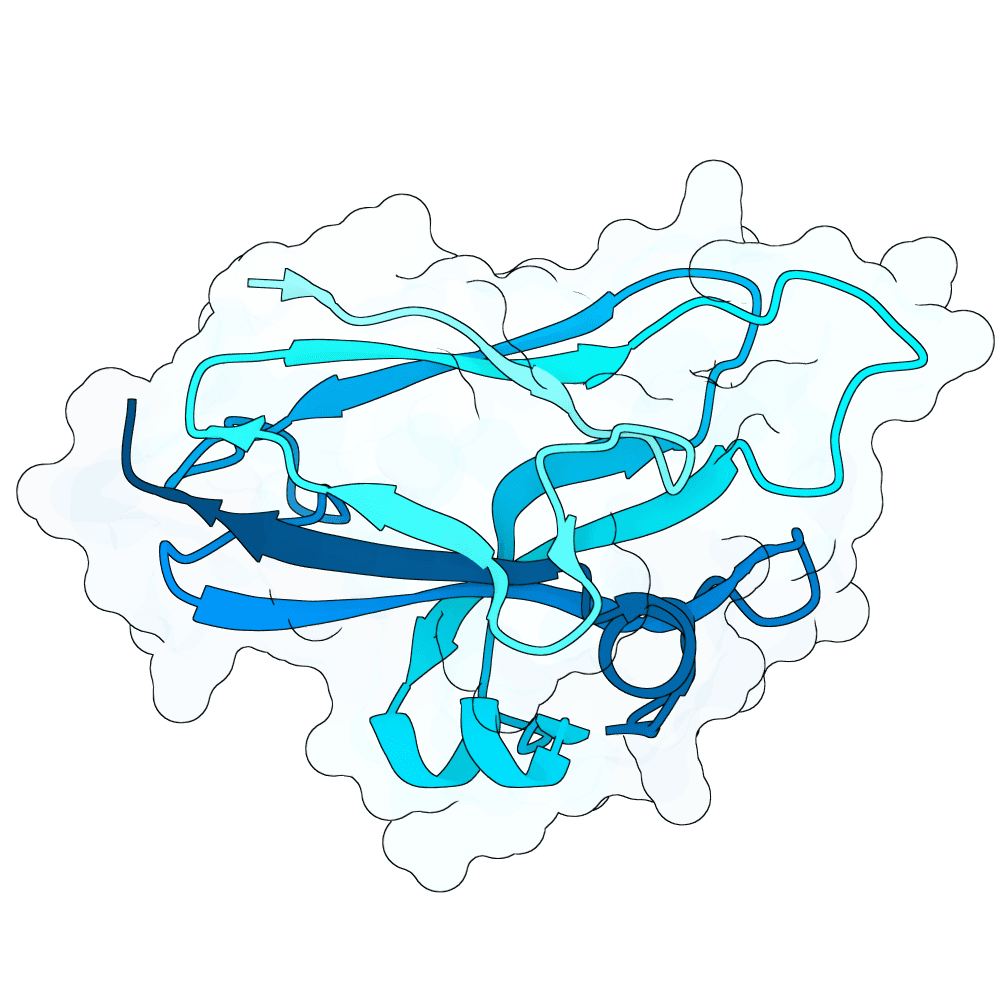
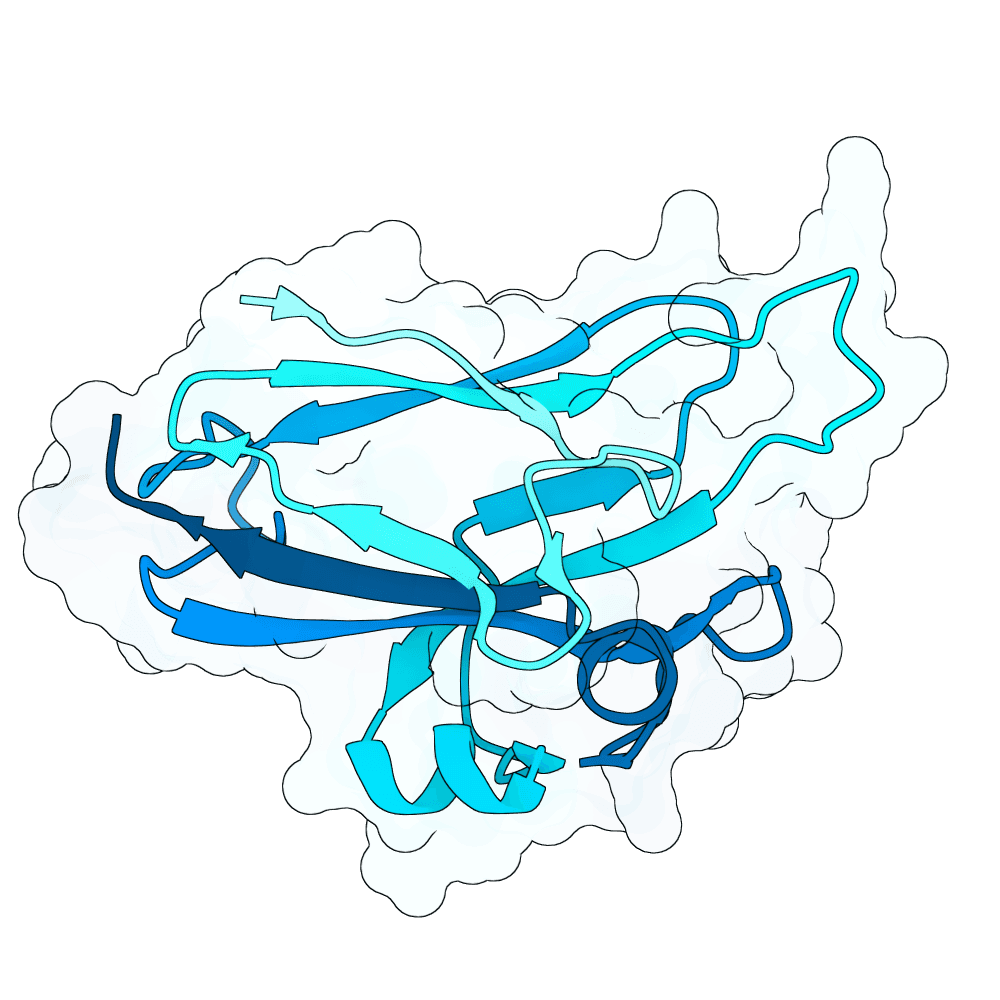
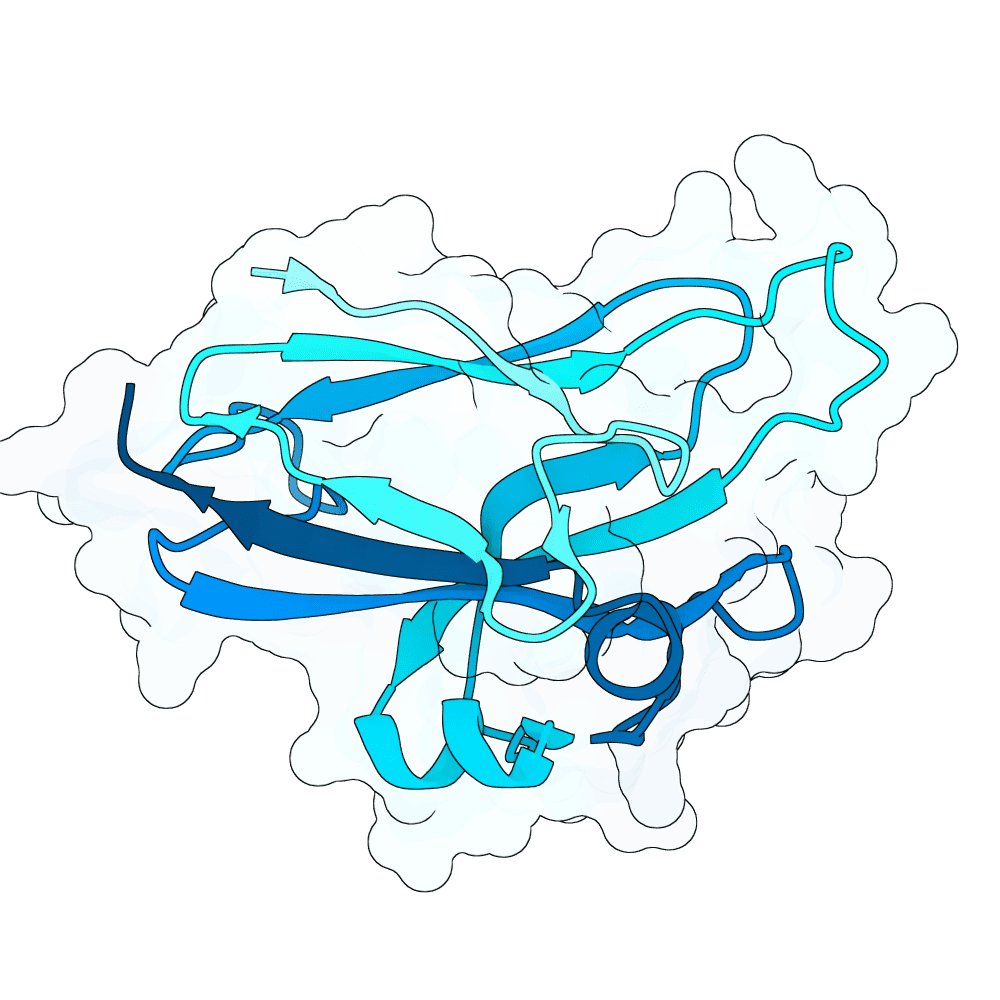
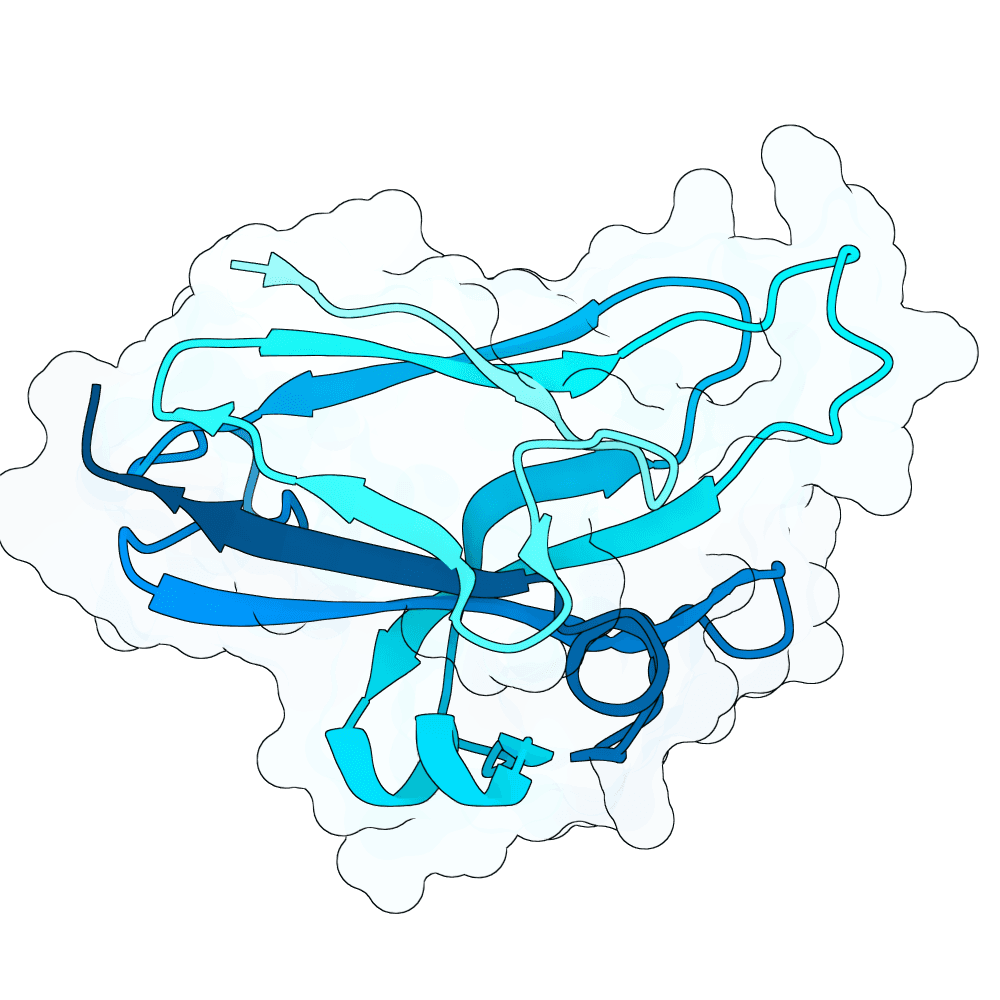
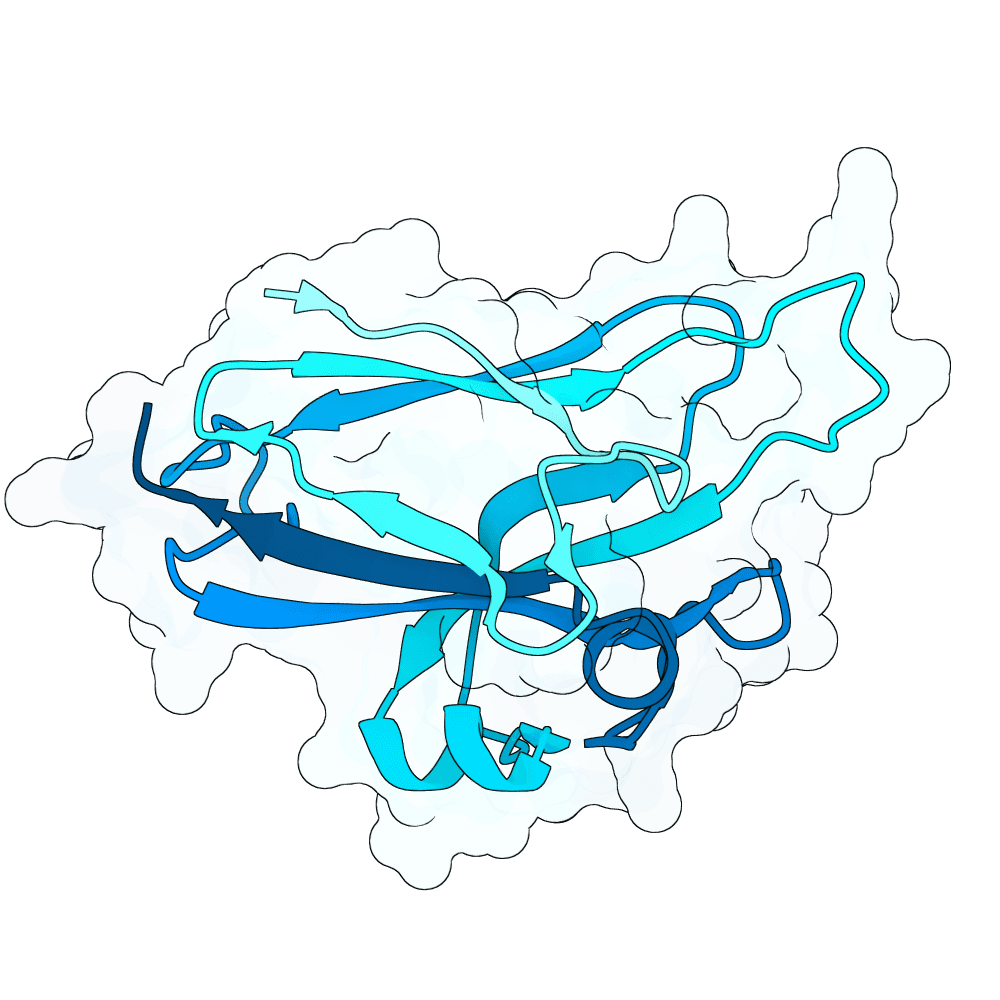
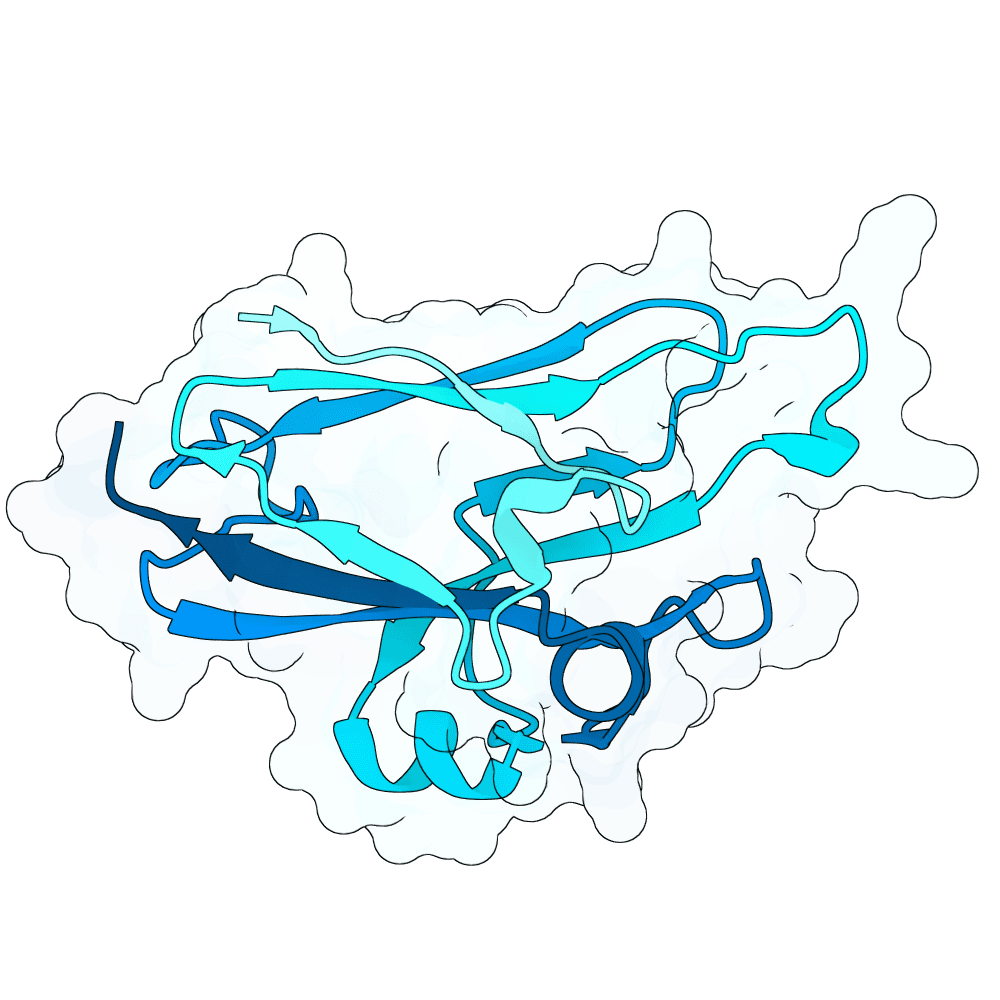
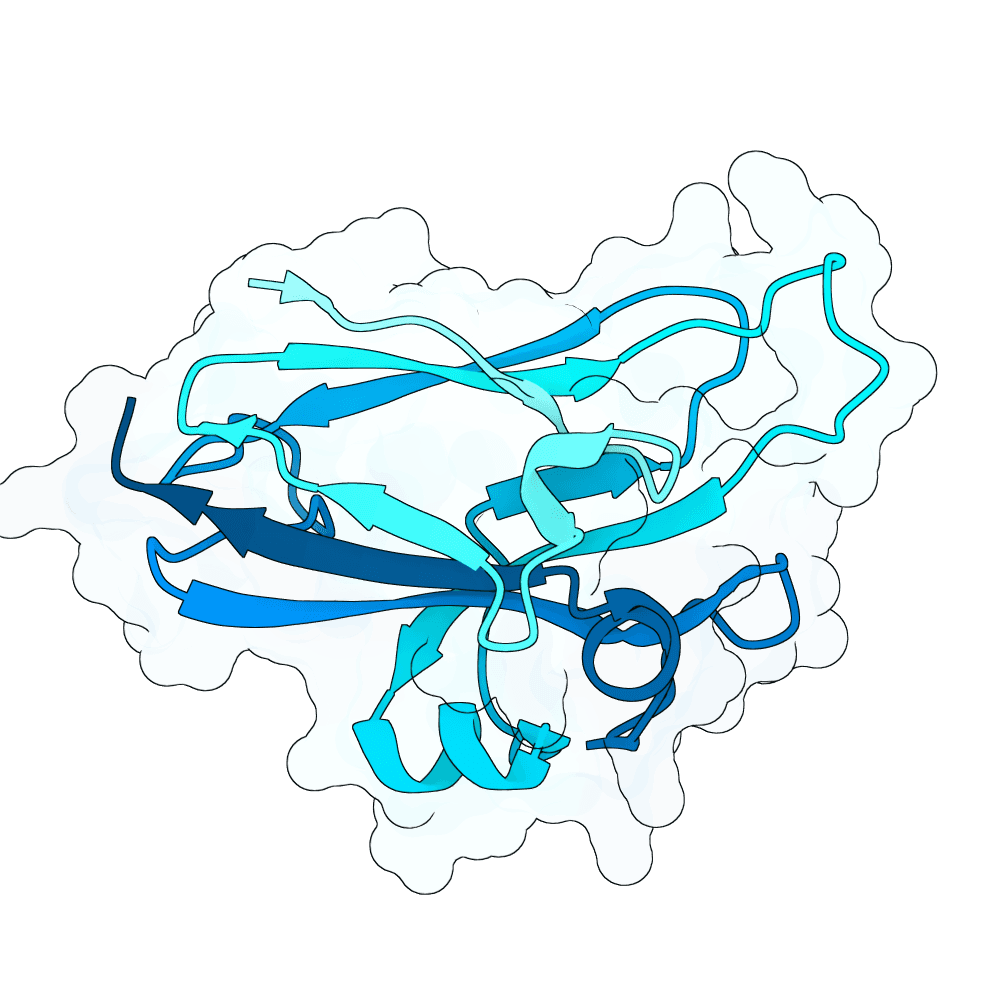
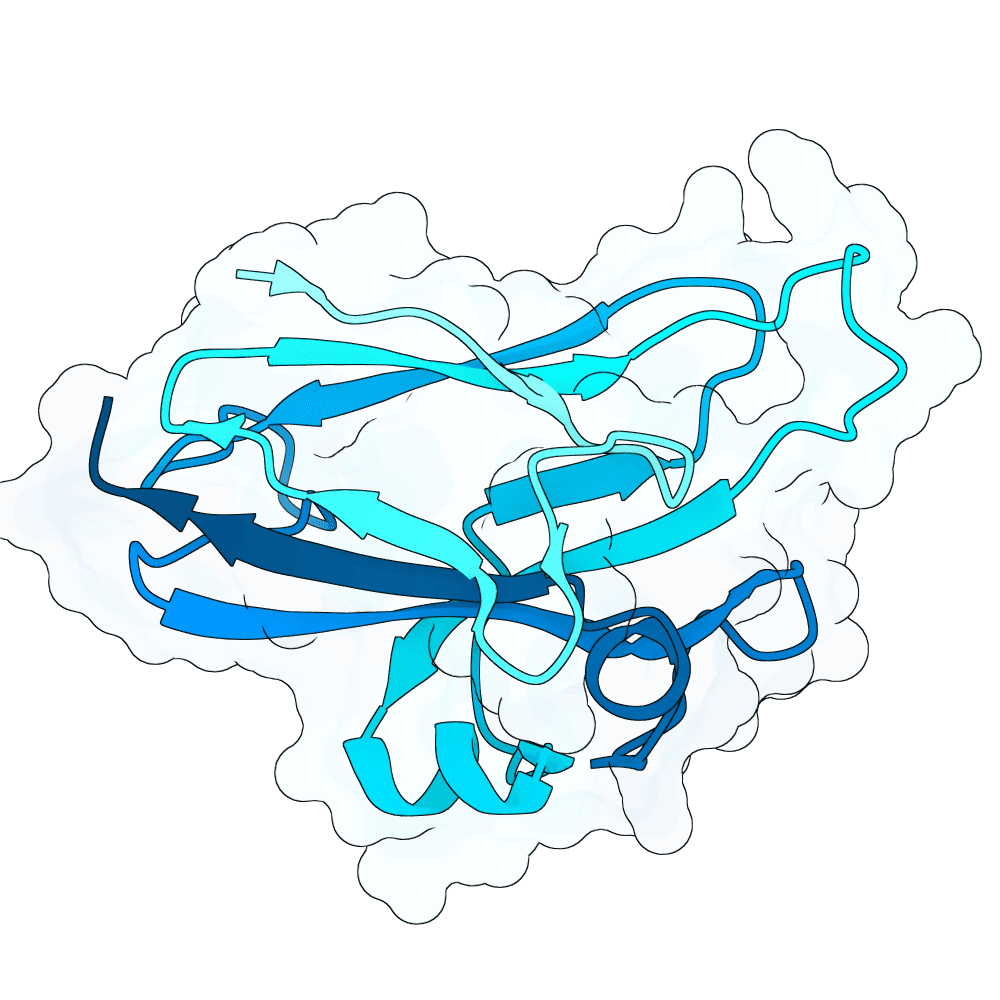
Description
All proteins were designed using an iterative process of using ProteinMPNN to generate variants of Ephrin B-2, a known substrate
Then, iterative quantitative analysis using AlphaFold and IPSAE to score proteins and analyze features of high-scoring sequences was used to filter and create new sequences, which were eventually submitted
According to AF3 and ipSAE, final protein sequences gave ipSAE scores of above 0.80 (but it appears that Boltz2 scores are lower)