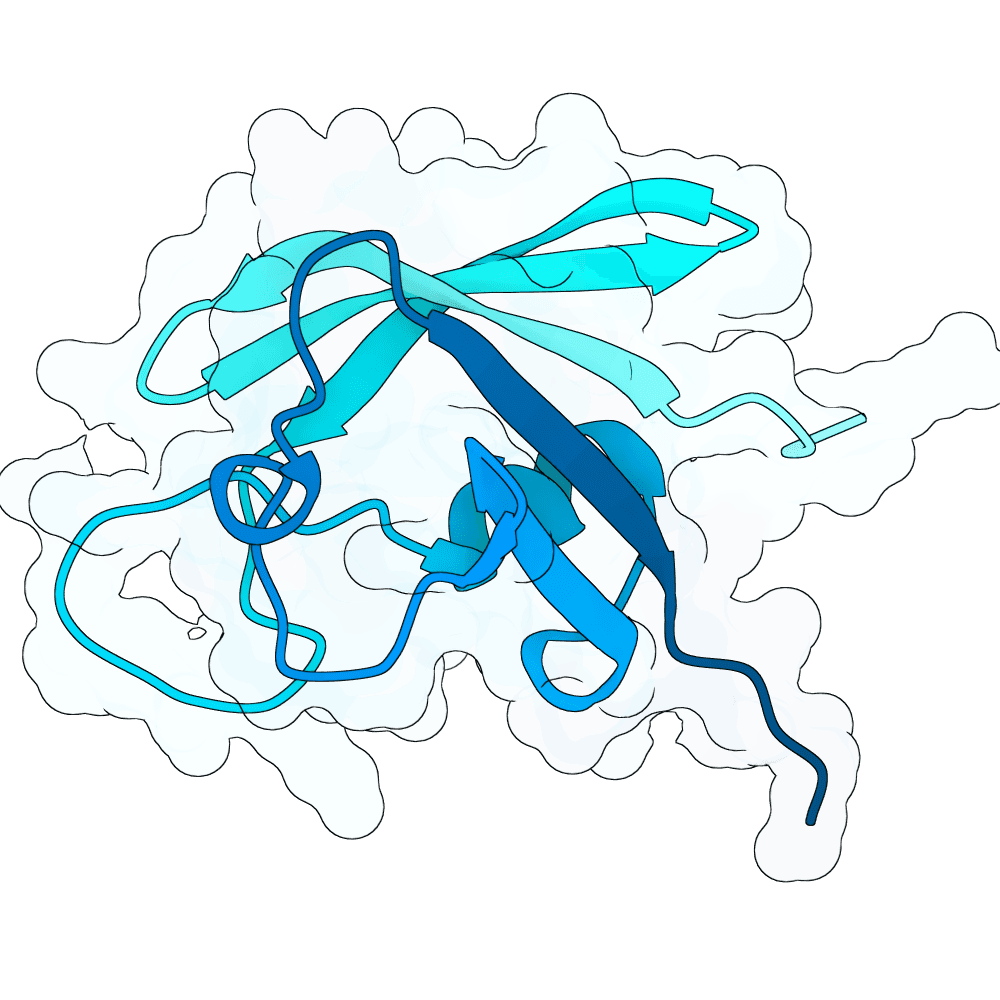
Description
The designed sequences were generated using the Kosmos AI Scientist from Edison Scientific. Kosmos was provided a prompt describing the competition rules, the protein structure 2VSM.pdb, Adaptyv's description of Surface Plasmon Resonance from its website, and a request to return 100 ranked candidate designs. It produced a .csv file with 100 ranked amino acid sequences and various metrics it derived. The submitted sequences are the top-ranked sequences from this list that passed the submission validity filters, i.e., excluding those that are too similar to published proteins.
The complete Kosmos prompt, intermediate .ipynb notebooks, output .csv file, generated report, and link to its shared results are provided at https://github.com/gitter-lab/adaptyvbio-nipah.