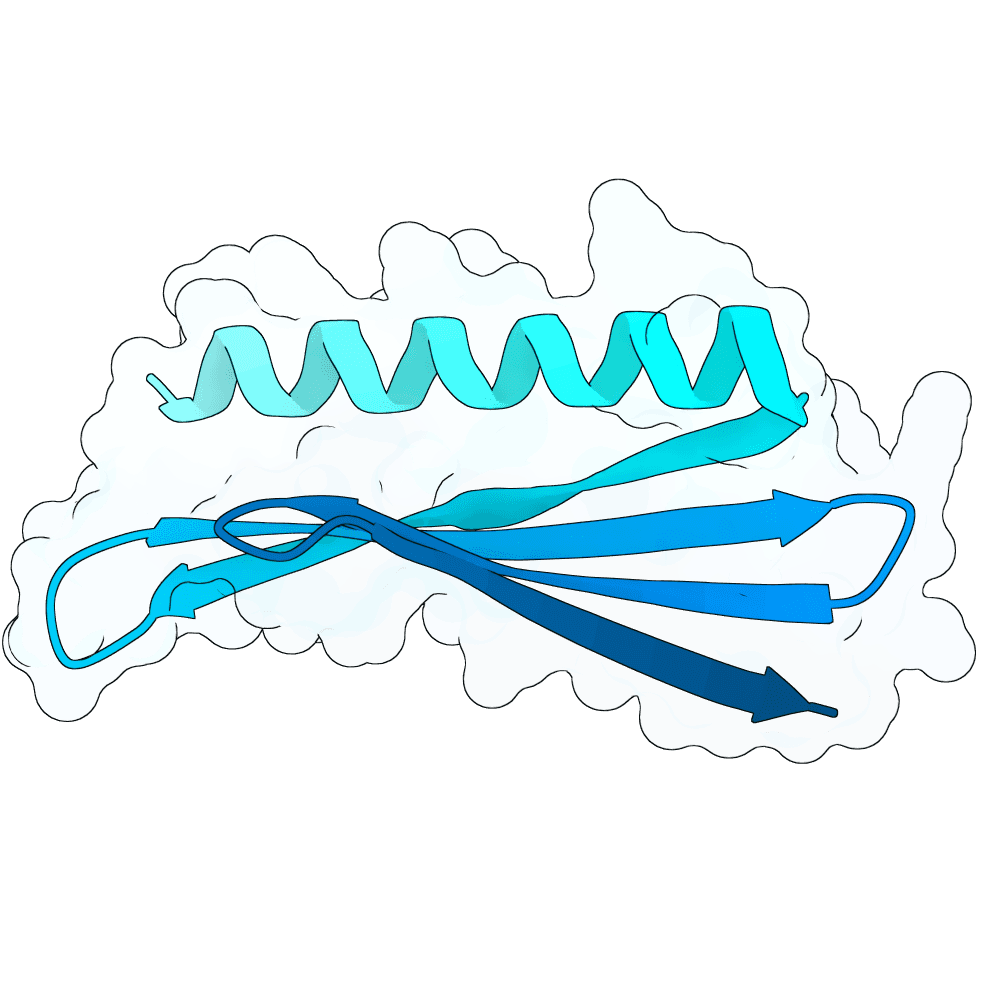
Description
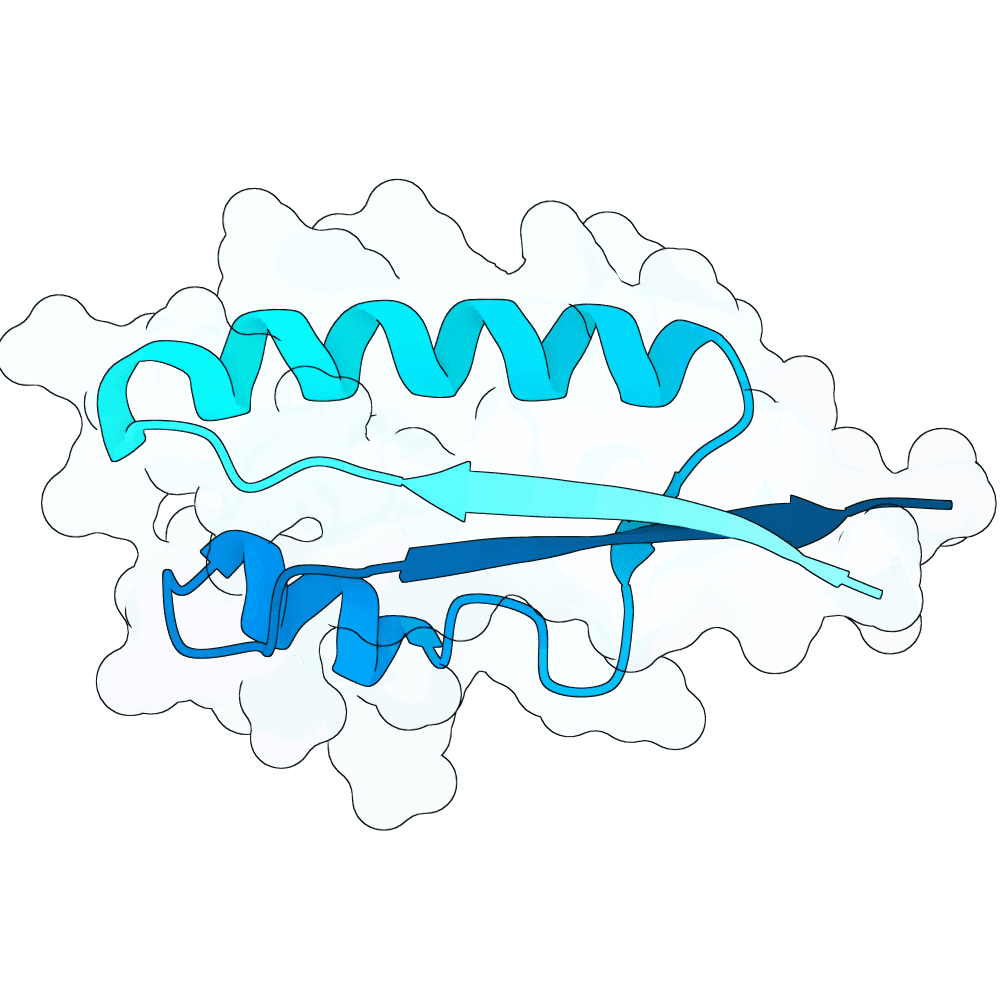
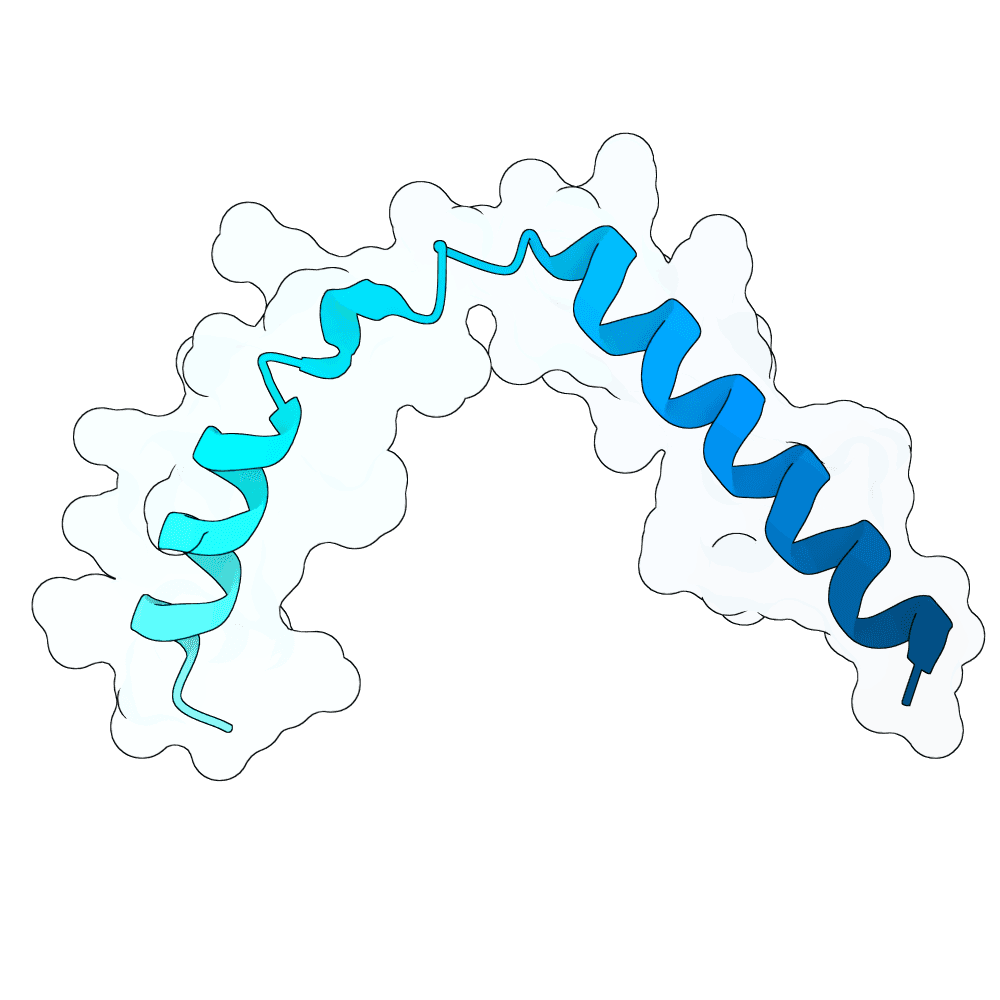
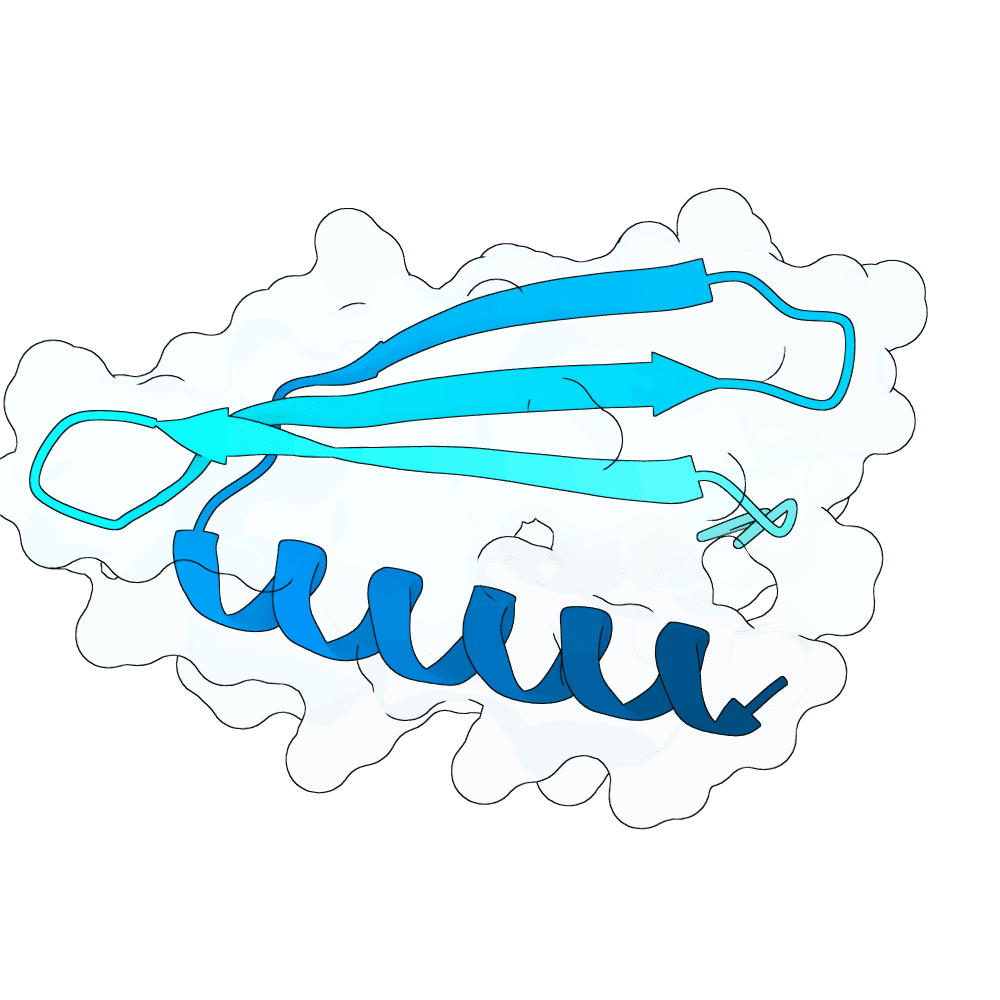
Starting from a neutralizing antibody–bound G protein complex (PDB 8JA5), I identified five sets of residues forming contacts with the G protein across five distinct loops. Keeping these interacting residues fixed, I used RFdiffusion to generate binders that preserve these contacts. Binder sequences were then designed with ProteinMPNN and folded with AlphaFold3. I filtered candidates using pLDDT, iPTM, global RMSD, and designed-interface RMSD to select stable, high-confidence designs.