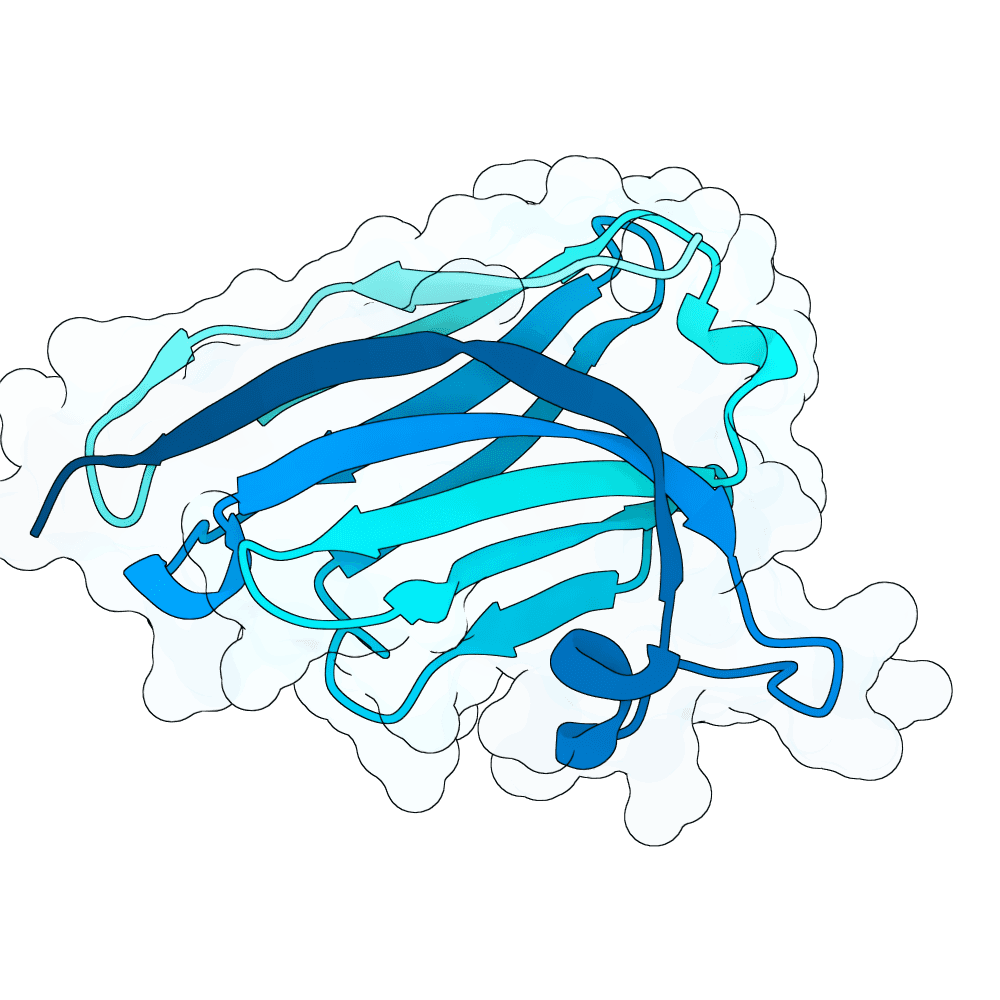
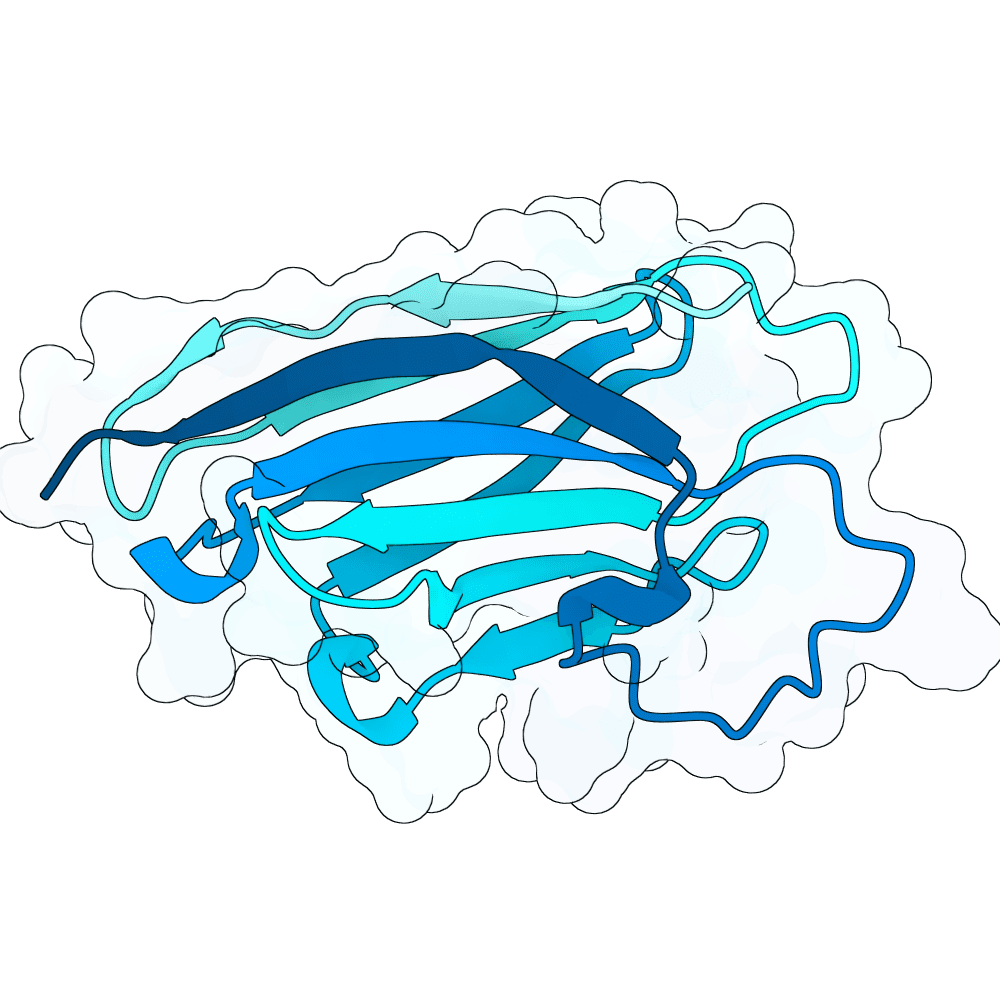
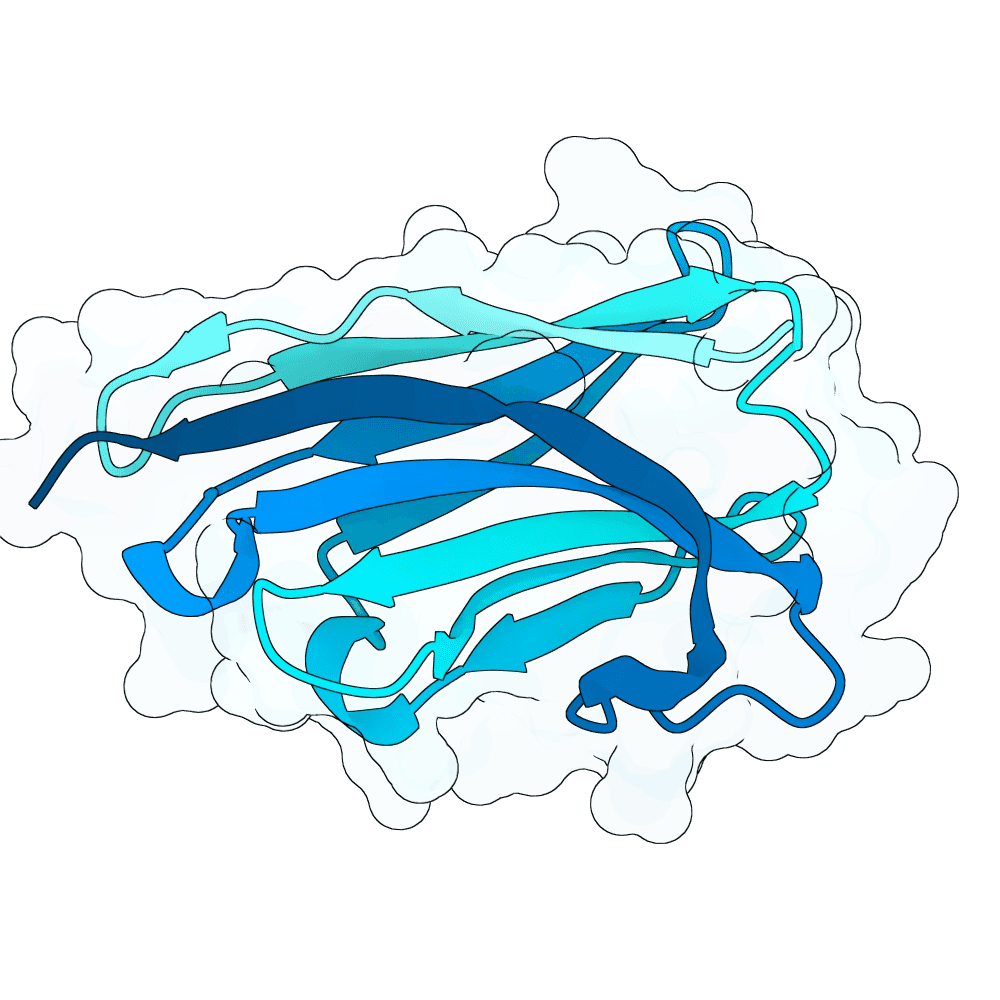
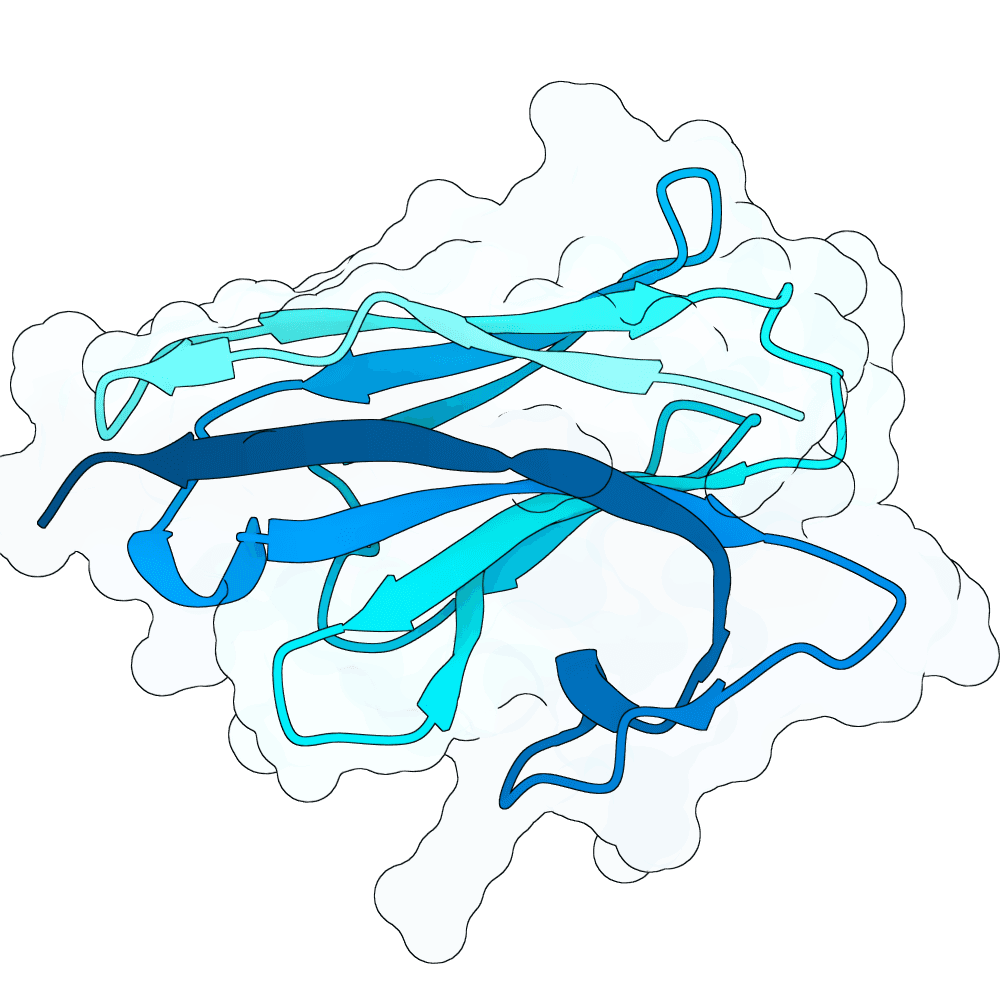
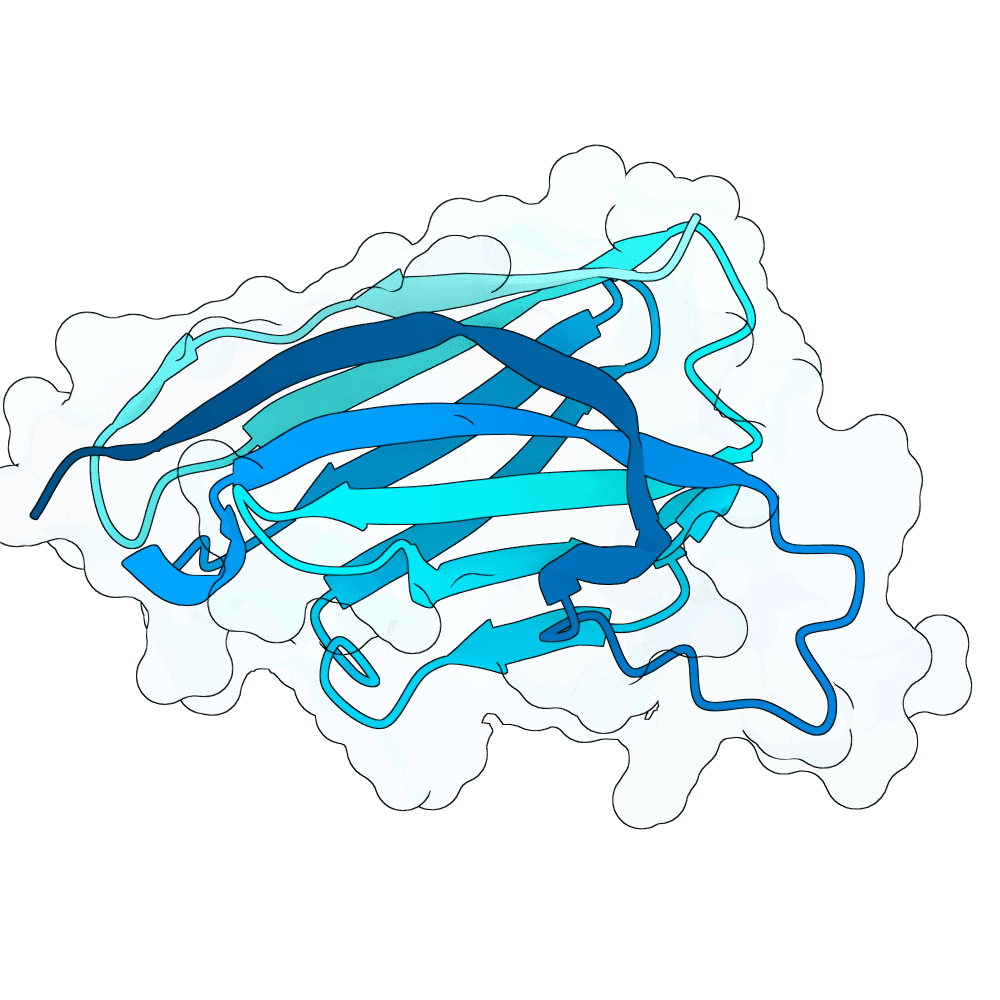
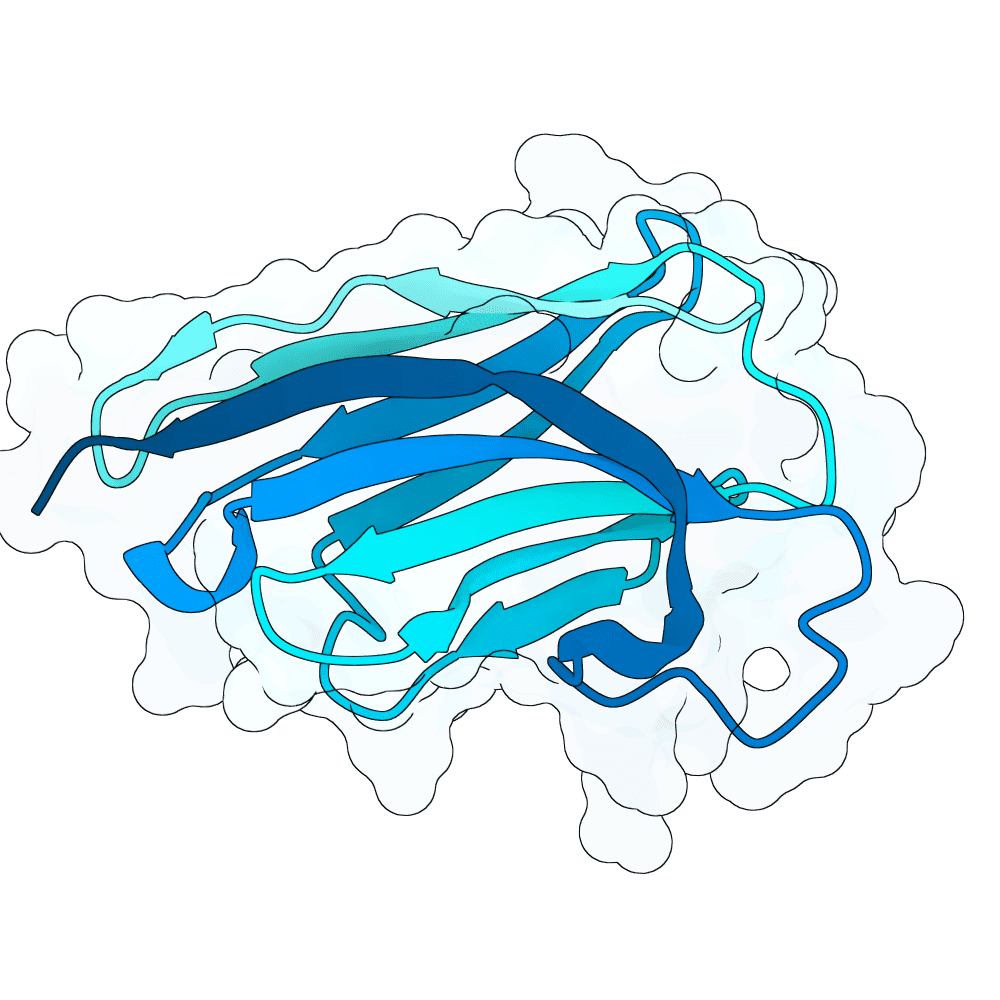
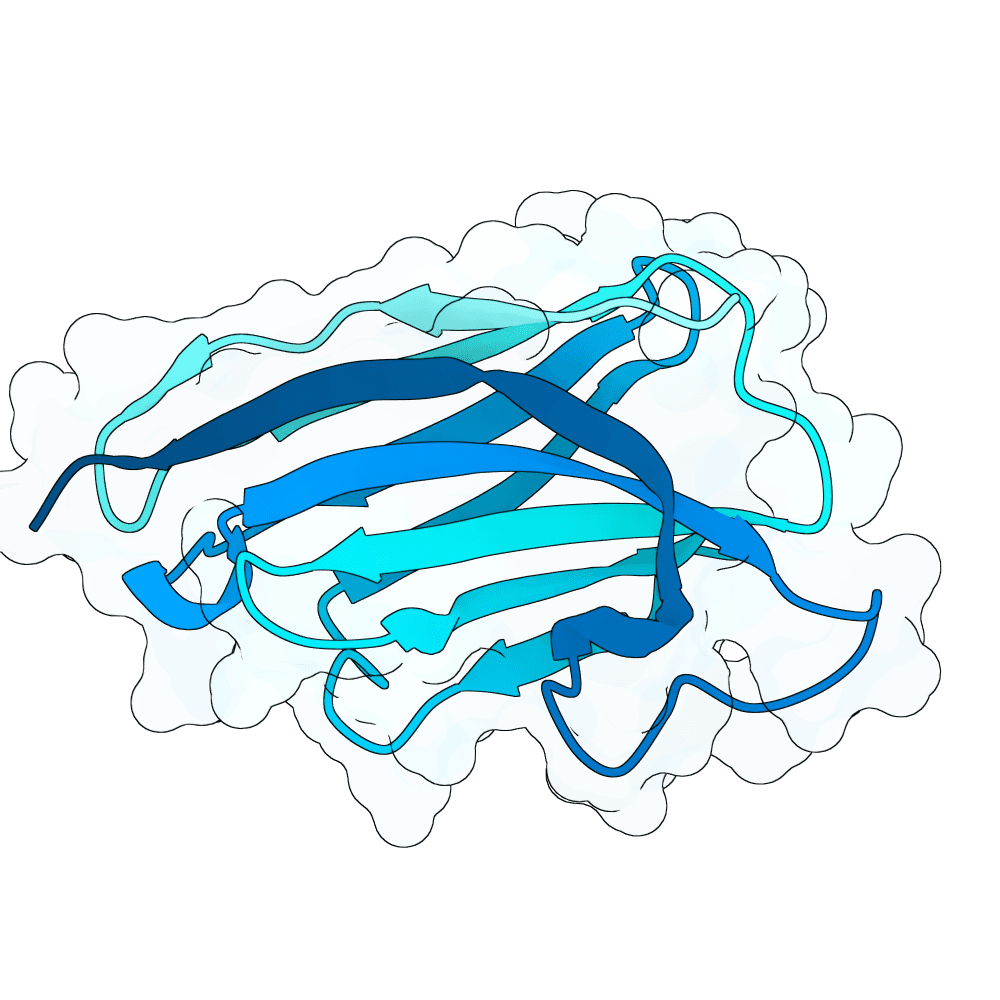
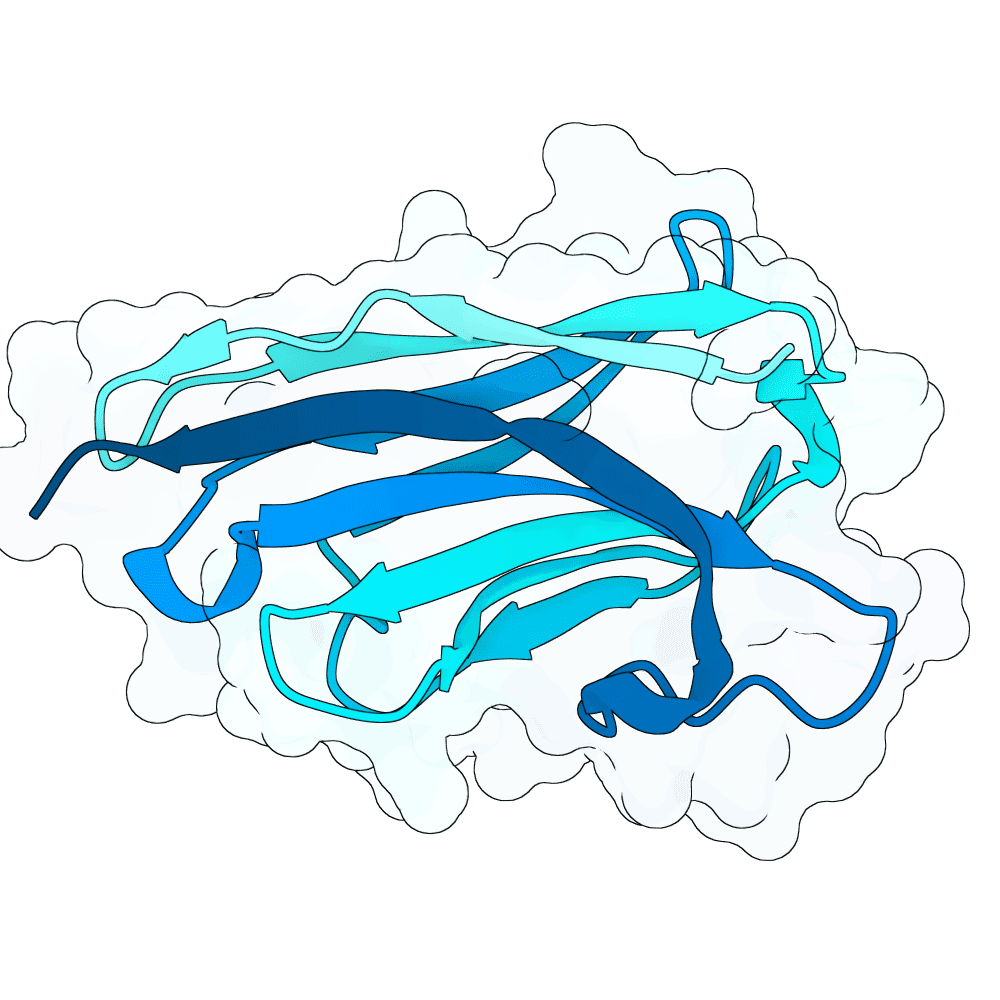
Description
We strongly advise the immediate manual selection and thorough experimental validation of our curated set of ten novel nanobody sequences. These sequences have been meticulously designed using clinically proven structural frameworks sourced from leading commercial and clinical-stage nanobody therapeutics, ensuring inherent stability and favorable pharmacological properties suitable for real-world therapeutic use. The engineering process focused specifically on refining the three Complementarity-Determining Regions (CDRs)—the essential binding loops—to achieve highly specific, strong antigen recognition that goes beyond conventional in silico homology-based screening.
Importantly, our design approach combined the generative capacity of Boltz-Gen for broad sequence exploration with our in-house structure prediction tool, DynaHelix. DynaHelix is uniquely tailored to improve prediction accuracy by automatically sampling multiple potential binding interfaces during modeling, overcoming the constraints of single static structure predictions and helping to identify the most favorable binding pose. Although the resulting CDR-antigen interaction interfaces may show lower scores in standard metrics such as ipTM or PAE—an anticipated outcome when designing sequences not present in existing structural databases—we assert that the robustness of our structure-aware validation and multi-model refinement supports their candidacy. This approach prioritizes functional viability and therapeutic relevance over high prediction scores alone.
As a result, these ten structure-informed candidates present innovative binding mechanisms and structural strategies that merit expert evaluation. They represent a substantial strategic opportunity to advance the development of next-generation nanobody-based therapeutics.