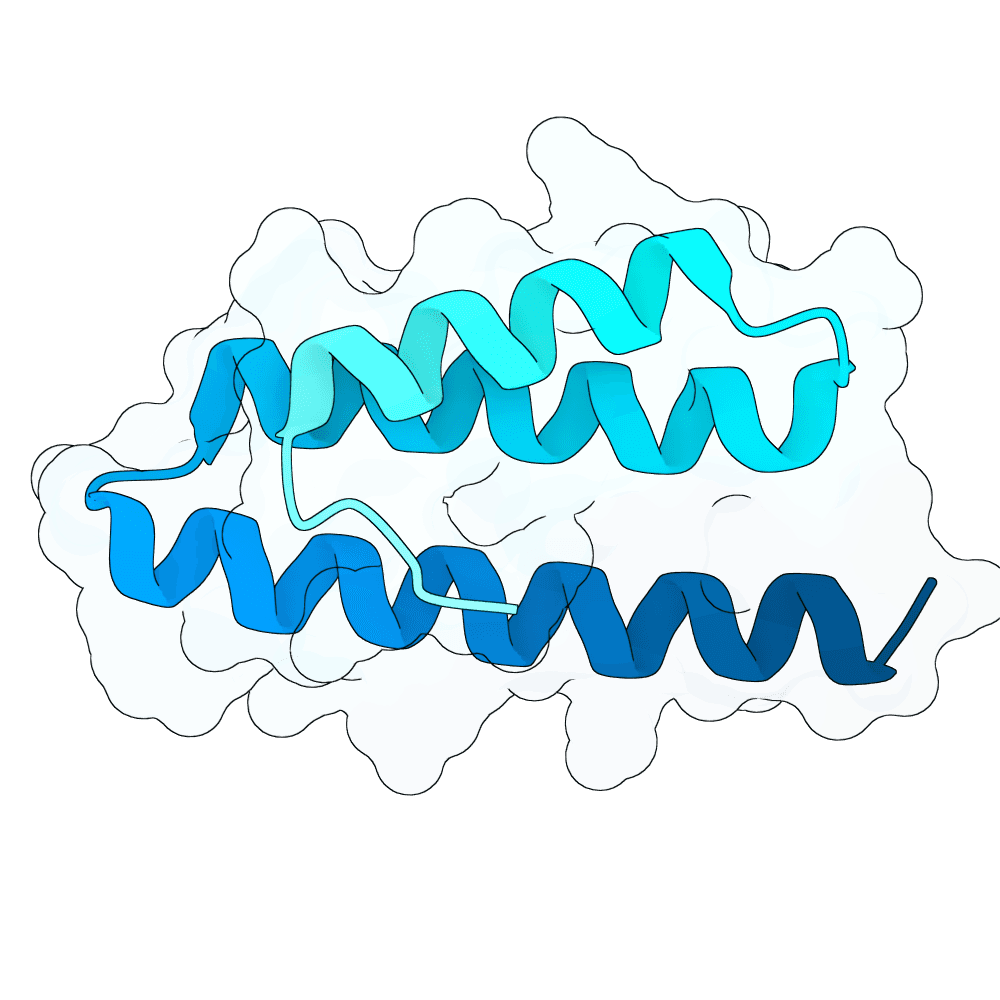
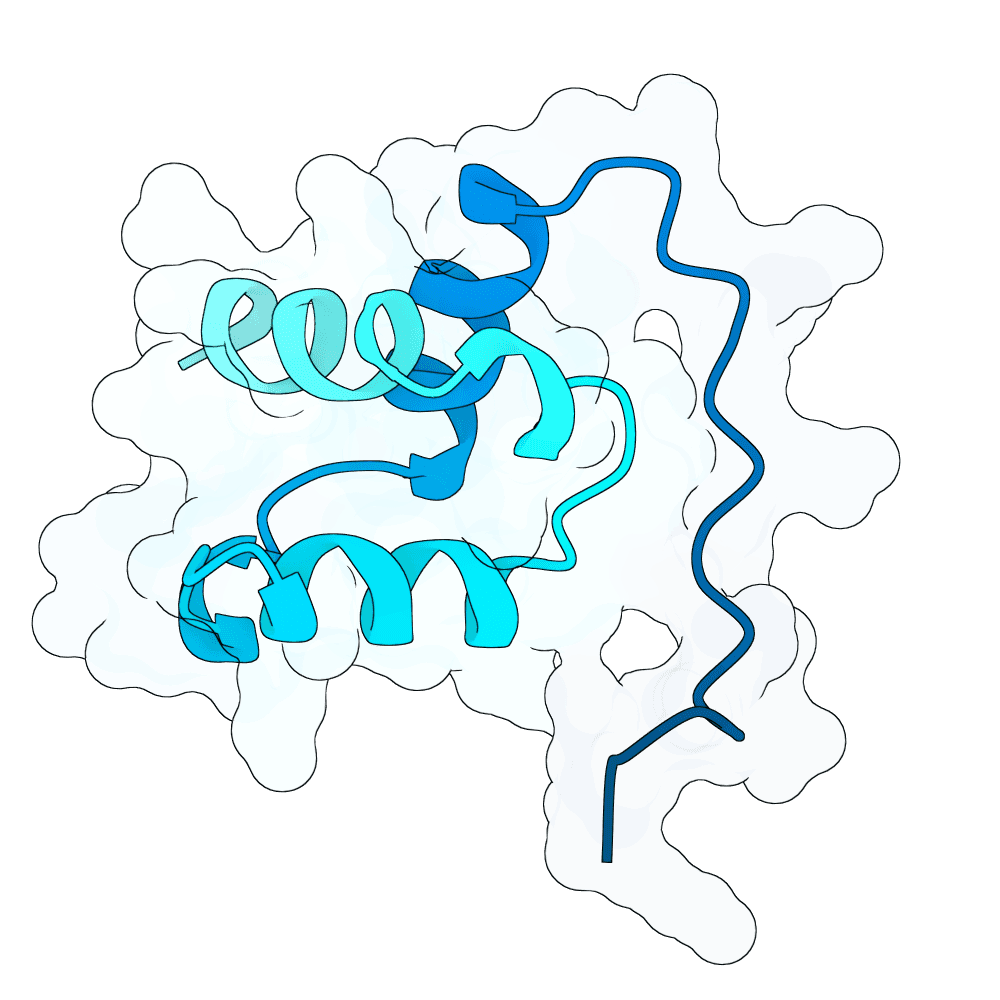Description
The binders were designed using adjusted versions of ESM3, LaProteina, APM, BindCraft, latentx and DPLM models. The final sequences were filtered by protT5 model. The work was jointly developed by our group (Yichen Zhou, Amir Karimi, Vincent Pauline, and Stefan Bauer), in collaboration with Emirhan Yagmur, Christoph Gruber, Moritz Haffner, Barbara Tremmel, and Ariane Linda Krus