Proteins (5)
id: rapid-bison-thorn
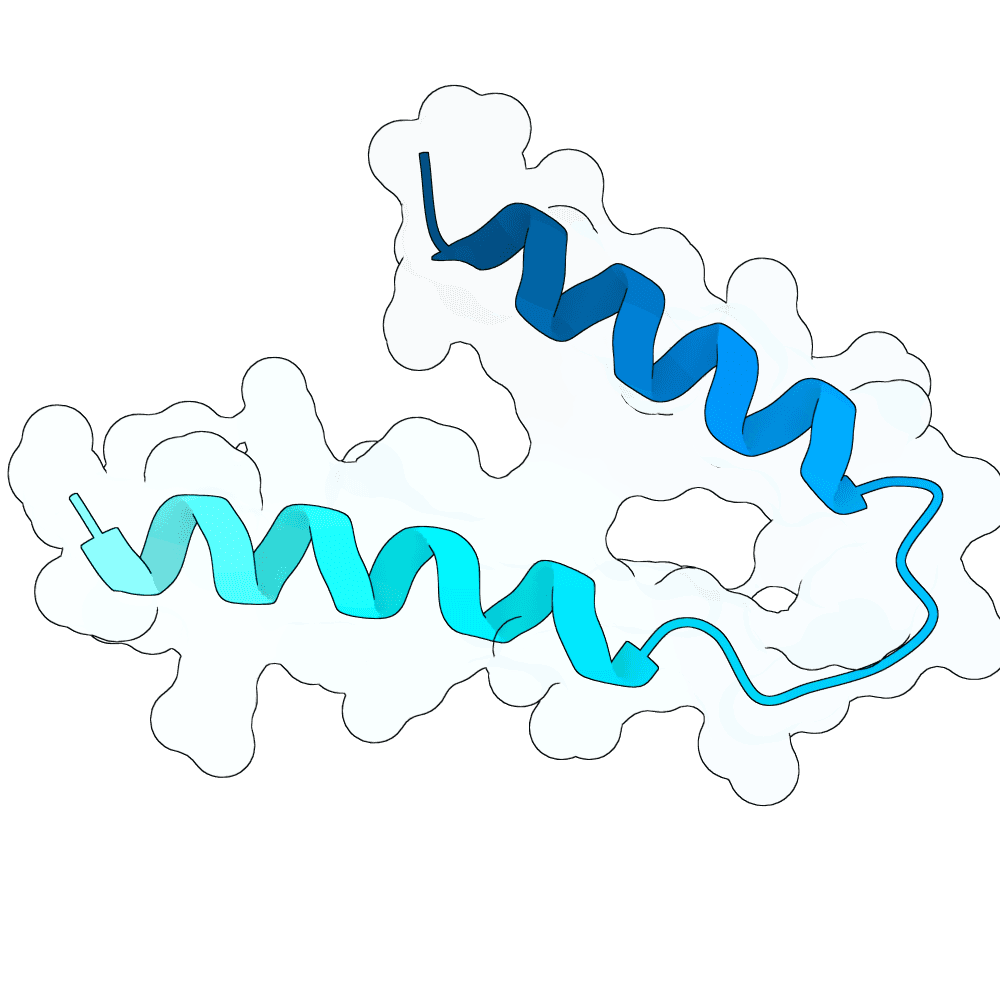
Nipah Virus Glycoprotein G
0.24
46.77
--
5.1 kDa
43
id: solid-goat-birch
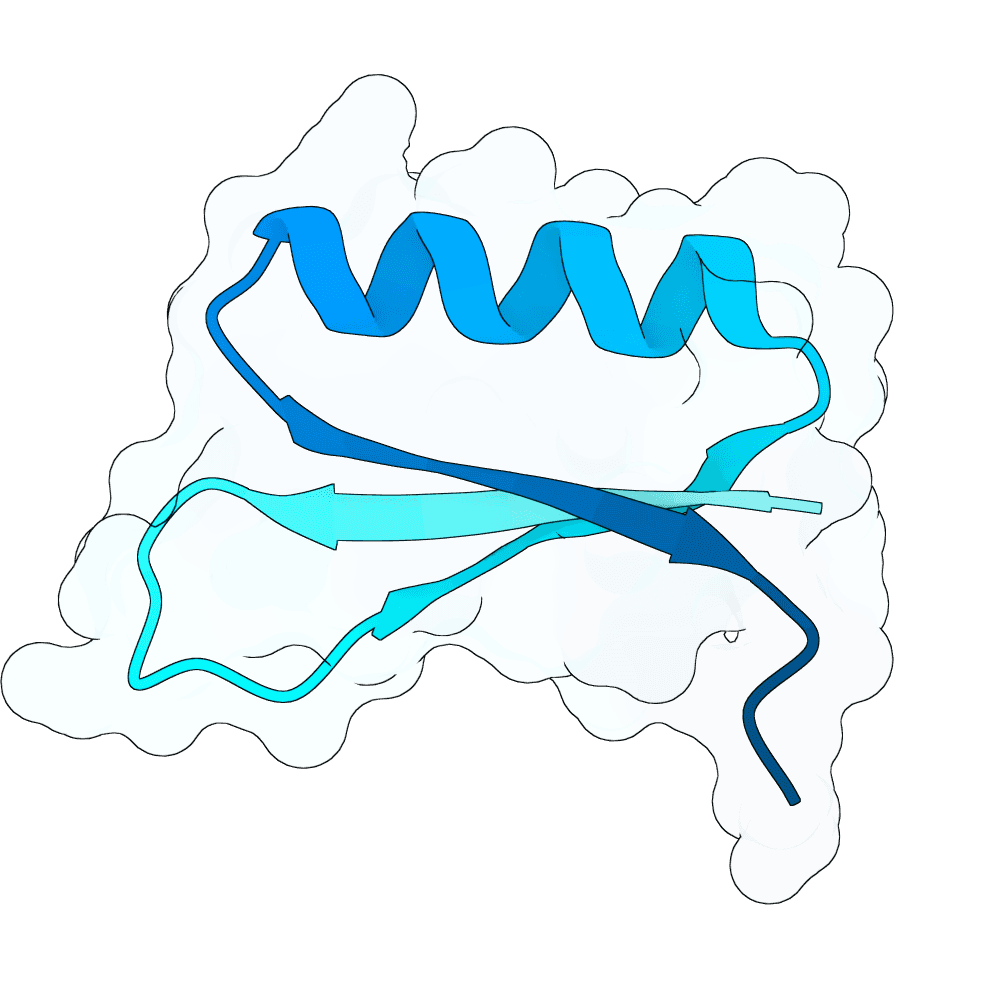
Nipah Virus Glycoprotein G
0.81
81.97
--
5.6 kDa
48
id: soft-swan-cloud

Nipah Virus Glycoprotein G
0.23
47.17
--
5.7 kDa
47
id: vast-moth-snow
Nipah Virus Glycoprotein G
0.22
70.94
--
4.1 kDa
34
id: strong-bear-willow
Nipah Virus Glycoprotein G
0.61
76.40
--
4.7 kDa
41