Proteins (10)
id: pale-kiwi-fern
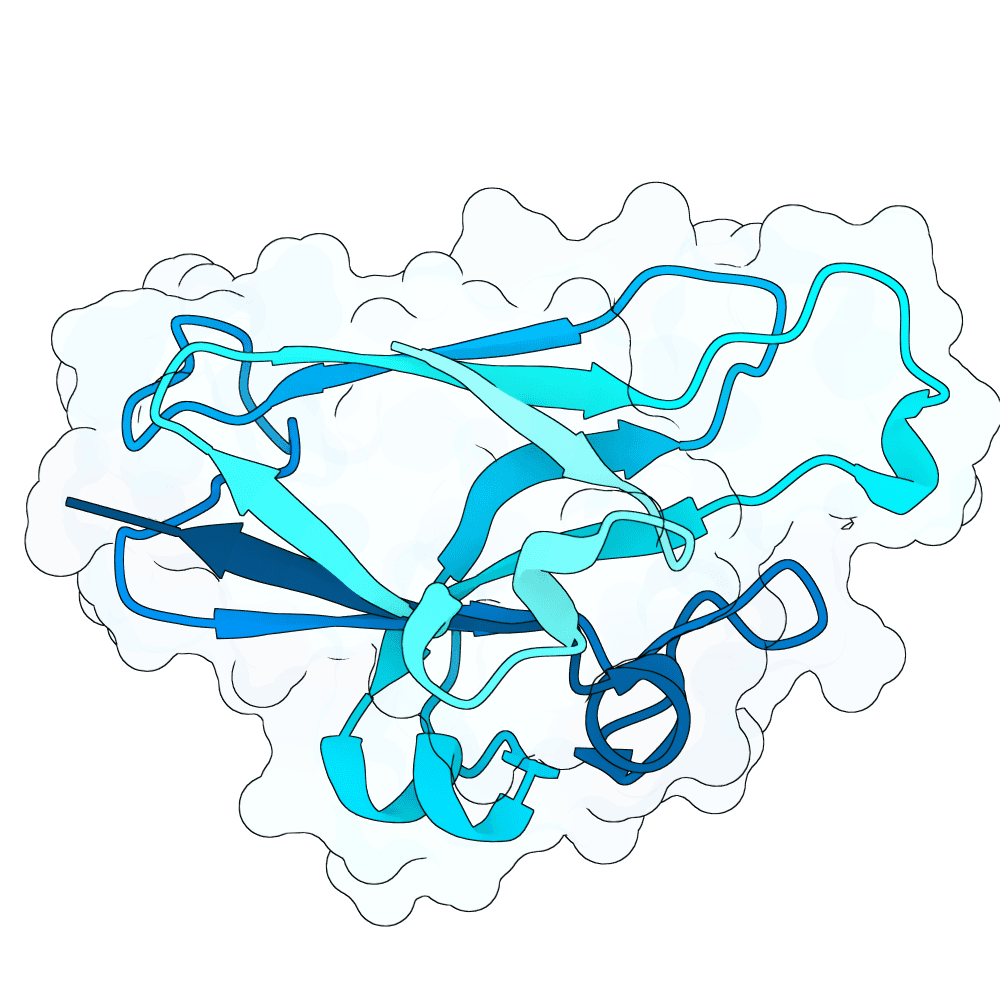
Nipah Virus Glycoprotein G
0.11
78.16
--
15.6 kDa
134
id: steady-owl-ash

Nipah Virus Glycoprotein G
0.75
77.28
--
14.5 kDa
125
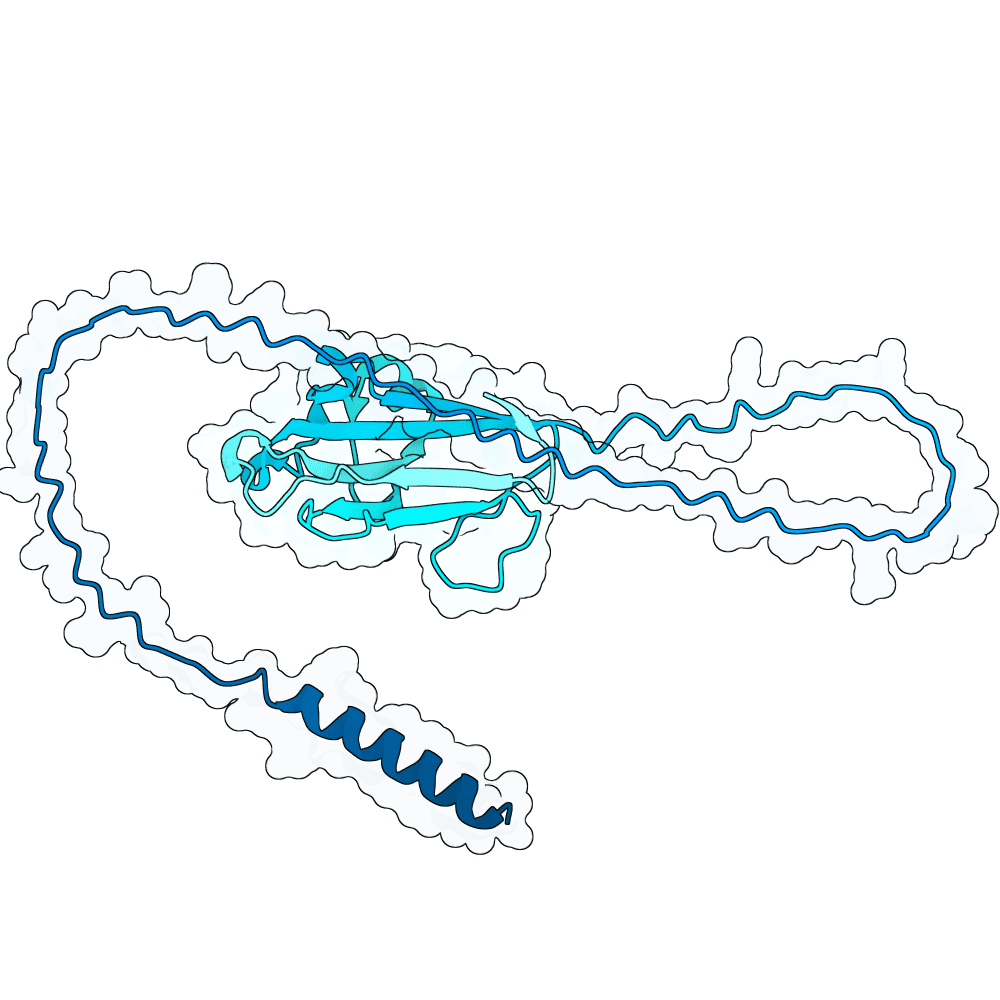
id: frozen-lion-opal
Nipah Virus Glycoprotein G
0.13
75.94
--
25.2 kDa
229
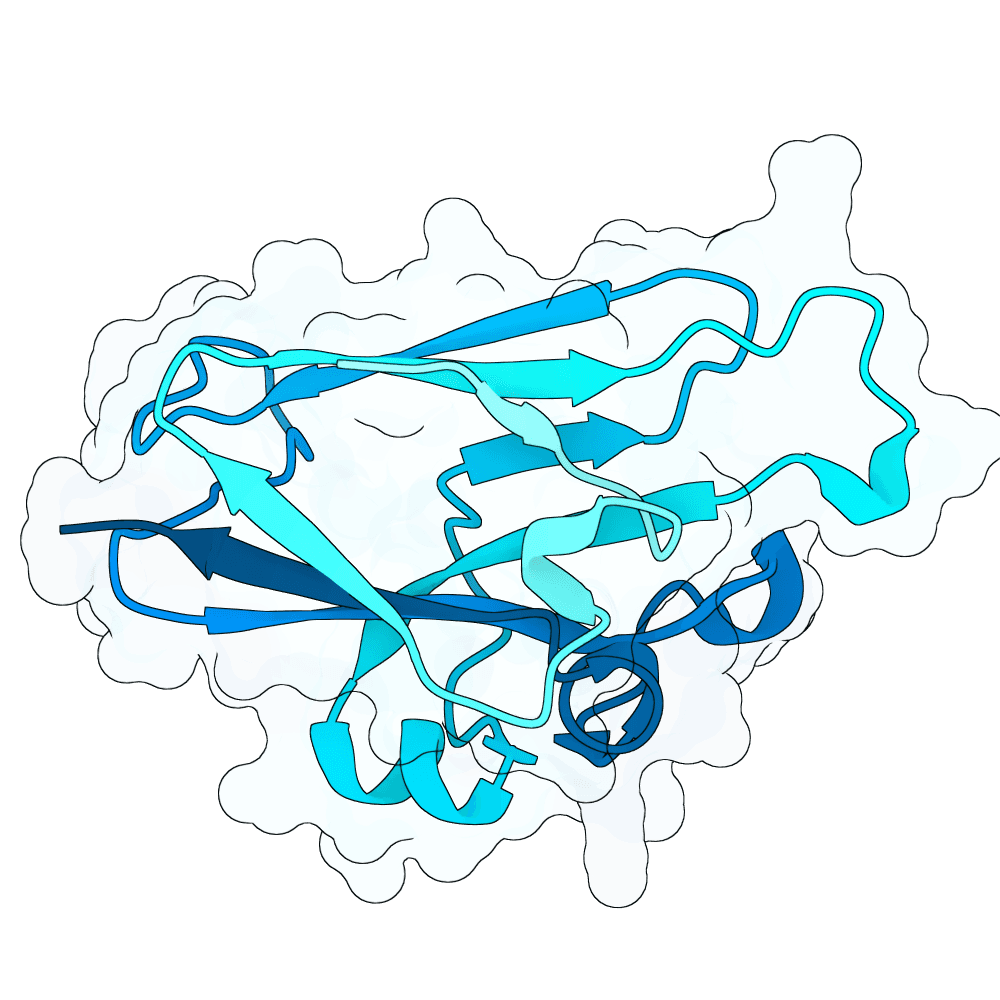
id: vast-fox-birch
Nipah Virus Glycoprotein G
0.07
81.66
--
15.5 kDa
137
id: dark-lynx-stone
Nipah Virus Glycoprotein G
0.36
81.52
--
15.1 kDa
132
id: jade-deer-oak
Nipah Virus Glycoprotein G
0.27
76.15
--
14.8 kDa
130
id: gentle-swan-ember
Nipah Virus Glycoprotein G
0.38
80.87
--
15.0 kDa
135
id: shy-lynx-wave
Nipah Virus Glycoprotein G
0.00
78.72
--
15.6 kDa
132
id: crimson-deer-quartz
Nipah Virus Glycoprotein G
0.00
83.15
--
15.3 kDa
134
id: rapid-deer-lotus
Nipah Virus Glycoprotein G
0.13
83.48
--
15.6 kDa
135