Proteins (10)
id: bright-bee-wave
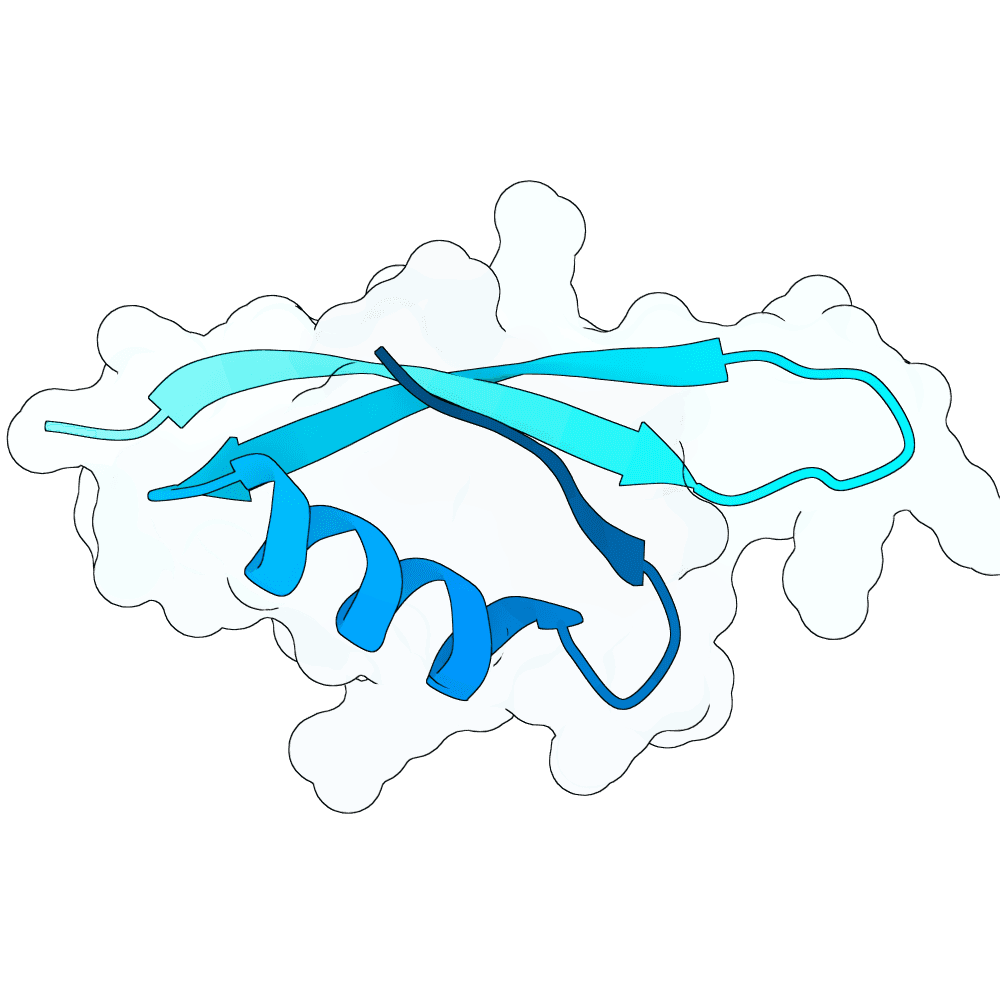
Nipah Virus Glycoprotein G
0.58
80.99
--
5.5 kDa
49
id: deep-raven-granite
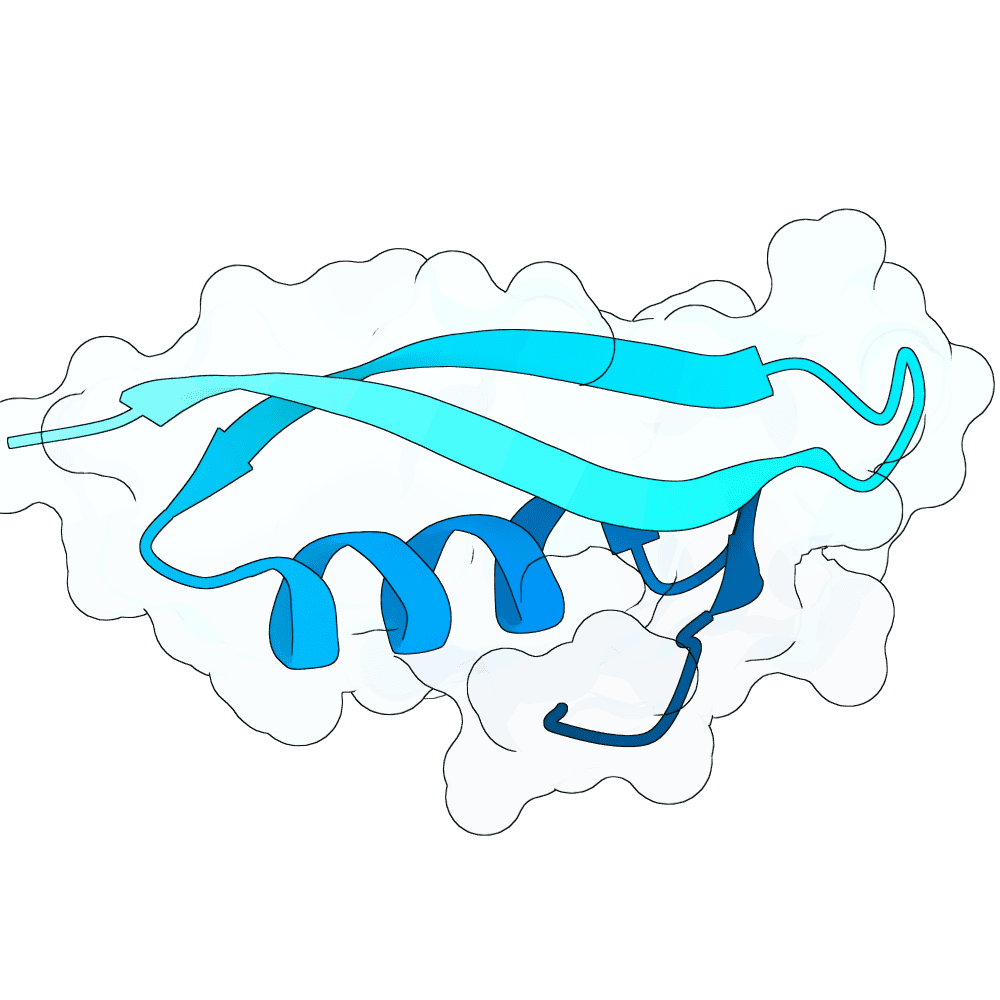
Nipah Virus Glycoprotein G
0.75
75.57
--
5.6 kDa
49
id: noble-toad-ember
Nipah Virus Glycoprotein G
0.83
84.54
--
4.3 kDa
41
id: rough-quail-willow
Nipah Virus Glycoprotein G
0.84
52.86
--
4.5 kDa
42
id: violet-ox-lotus

Nipah Virus Glycoprotein G
0.55
90.00
--
5.9 kDa
51
id: deep-dove-frost
Nipah Virus Glycoprotein G
0.39
46.03
--
6.0 kDa
51
id: swift-falcon-wave
Nipah Virus Glycoprotein G
0.81
80.12
--
4.8 kDa
42
id: quick-heron-dust
Nipah Virus Glycoprotein G
0.76
79.58
--
4.5 kDa
42
id: steady-cobra-quartz
Nipah Virus Glycoprotein G
0.69
77.45
--
4.5 kDa
41
id: hollow-falcon-opal
Nipah Virus Glycoprotein G
0.59
61.17
--
5.4 kDa
45